Abstract
In recent years, circular RNAs (circRNAs), a new class of RNA molecules characterized by their covalently closed circular structure, have become a new research paradigm in RNA biology. Many circRNAs are conserved among eukaryotes, localize in specific subcellular compartments, and play different biological roles. Accumulating evidence shows that circRNAs regulate a diversity of cellular processes by acting as miRNA sponges, anchors for circRNA binding proteins (cRBPs), transcriptional regulators, molecular scaffolds, and sources for translation of small proteins/peptides. The emergence of the biological functions of circRNAs has brought a new perspective to our understanding of cellular physiology and disease pathogenesis. Recent studies have shown that the expression of circRNAs is tissue- and cell type-specific and specifically regulated through development or disease progression, where they exert specific biological functions. However, the mechanisms underlying these remain largely unknown. A deeper understanding of how the specific expression of circRNAs is regulated to exert specific biological functions will enable the use of circRNA as a biomarker in clinical practice and the development of new therapeutic approaches. This review aims to summarize recent developments in circRNA biogenesis, functions, and molecular mechanisms. We also provide some specific circRNAs as examples to show their tissue-specific distribution and evaluate the possibility of applying circRNA technologies in molecular research and therapeutics.
Similar content being viewed by others
Facts
-
The number of circRNAs detected is significantly more than the number of genes.
-
The abundance of different circRNAs generated from the same gene or different genes is very different.
-
Many circRNAs are expressed at very low levels.
-
An exon may be shared by different circRNAs.
Questions
-
What splicing factors are controlling the biogenesis pathway associated with the abundance of different circRNAs generated from the same gene?
-
Are the low abundant circRNAs tissue-, cell type-, and developmental stage-specific or are they expressed at low abundance all the time?
-
Do these low abundant circRNAs affect cell activities?
-
For the circRNAs sharing the same exon(s), do they also share similar functions?
Introduction
CircRNAs, a novel class of non-coding RNA, were first identified about 40 years ago, which are widely distributed in fungi, protozoa, plants, mice, humans, etc. [1, 2]. The circRNAs have been considered as the products of splicing errors for a long time until recently the properties of circRNAs are successfully and extensively investigated with the new generation of RNA-seq and are demonstrated to play crucial roles in a variety of biological processes [3]. Soon after, circRNAs become one of the hottest topics in RNA biology [4]. CircRNAs have a unique covalently closed circular form and are generated from precursor mRNA by RNA polymerase II [4, 5]. They are abundant, distinct, highly conserved molecules which are expressed in tissue- and cell type-specific manners during developmental and disease stages [6,7,8]. In this review, we focus on the tissue-specific expression and functions of circRNAs. We also outline the probabilities of applying circRNA technologies in molecular therapy.
General features of circRNA
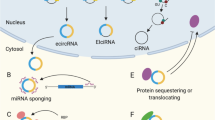
CircRNAs are covalently closed-loop molecules that lack 5′-caps and 3′-poly(A) tails and are resistant to RNA exonucleases-mediated degradation [9, 10]. The abundance of circRNAs is specific to cell types and tissues during development [7, 8]. In humans, high levels of circRNAs are observed in the brain, neural cells, heart, and lung [3, 11]. Some circRNAs are even more abundant than their linear counterparts [4, 12]. Although circRNAs are synthesized in the nucleus, they predominately localize in the cytoplasm. Most exonic circRNAs (ecircRNAs) are found to be transported to the cytoplasm by the nuclear export system [13]. The circRNAs are sorted according to their length by the nuclear export machinery and their localization is controlled by UAP56/URH49. UAP56 controls the export of long circRNAs (>1200nt) and URH49 is required for short circRNAs (<400nt) [14]. The m6A modification in circNSUN2 can facilitate its export [15]. In cytoplasm, circRNAs may function as microRNA sponges, interact with circRNA binding proteins (cRBPs), or serve as templates for translation. The majority of intronic circRNAs (ciRNAs) and exon-intron circRNAs (EIciRNAs) are retained in the nucleus [16]. The circular form of the introns of C9ORF72 transcripts can be exported to the cytoplasm and serve as the translation template [17].
Biogenesis and classification of circRNAs
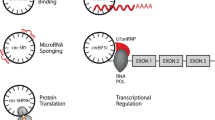
CircRNAs are generated from precursor mRNAs by RNA polymerase II, through which the 3’-end splice site is linked with the upstream 5’-end splice site to form a closed-loop, known as back-splicing [5]. There are three models of circRNA loop formation based on circRNA compositions: intron pairing-driven, RNA-binding protein (RBP)-driven, and lariat-driven circularization (Fig. 1) [18]. According to their genomic origin and cycling mechanism, circRNAs are divided into ecircRNAs, ciRNAs, and EIciRNAs [8, 16]. The ecircRNAs are mainly derived from single or multiple exons of pre-mRNA [19]. The ciRNAs are derived from introns by spliceosome-mediated splicing [20]. EIciRNAs consist of both exonic and intronic sequences [21]. RBPs play roles in circRNA biogenesis, while adenosine deaminase acting on RNA (ADARs) participates in the formation of circRNAs through A-to-I substitution [12].
a Linear mRNA is derived by the conventional splicing process. b Intron pairing model of circRNAs biogenesis; The 3’ splicing donor site in the exon combines with the 5’ splicing acceptor site in the upstream intron that forms a circRNA or ecircRNA by removing the introns between the exons. c Intron pairing-driven circularization; reverse complementary sequences (Alu sequences) direct insertion and generate an EIciRNA or ecircRNA through intron removed or retained. d RBP-induced cyclization: RBPs assist the back-splicing event, in which the introns are removed to form circRNA. e ciRNA formation; ciRNAs are derived from lariat introns. f circRNAs can serve as miRNA sponge. g circRNAs can interact with cRBPs and alter the functions of the proteins. h circRNAs can encode peptides and proteins. i ciRNAs can directly interact with transcription complexes and thus regulate parental gene expression at the transcriptional and translational levels.
Current approaches for circRNA detection and quantification
Different methodologies are employed to identify circRNAs to elucidate their functions [22]. The predictive tools, including circRNA_finder, find_circ, CIRCexplorer, CIRI, and MapSplice, determine circRNAs by aligning the reads of RNA-seq with a reference or pseudo-reference genome to extract reads that propagate to back-splicing junctions [18, 23]. The current online databases (CircRNADb, Circ2Traits, CircInteractome, CircBank, and CircNet, etc.) provide the basic information and classification of circRNAs, predictive analysis of circRNAs interactions, the relationship between circRNA and disease, primer design, and siRNA design [24,25,26,27,28]. Commonly used techniques for genome-wide profiling and quantification of circRNAs include Ribo-RNA-seq, poly(A)-RNA-seq, RNase R-treated RNA-seq, Reads per million mapped reads, fragments per million mapped fragments, CIRCscore, and circular over linear ratio [5].
Northern blotting is the initial assay for circRNA validation [29]. However, due to the complexity and the low sensitivity of Northern blot, the most commonly used technique for circRNA detection and validation is RT-PCR or quantitative PCR with divergent primers flanking the back-splicing junctions. The copy numbers of circRNA can be determined with digital droplet PCR (ddPCR) [30]. Fluorescence in situ hybridization (FISH) is performed with specific probes to visualize the subcellular location of circRNAs [31, 32]. Identification of the circRNA-binding proteins can be achieved by circRNA-pulldown method [33,34,35].
Molecular mechanisms and functions of circular RNA
Accumulated evidence has provided a focus on the potential roles of circRNAs. The known functional mechanisms of circRNAs include sponging miRNAs, interacting with cRBPs, translating proteins, and regulating transcriptional or post-transcriptional gene expression (Fig. 1) [12].
microRNA sponge
CircRNAs can bind different types and numbers of microRNAs and negatively regulate microRNA activity by competing with mRNA-microRNA binding [36, 37]. The ciRS-7 has over 70 miR-7 binding sites and functions as miR-7 sponge [38]. CircRNAs have been shown to be involved in many pathological processes by sponging different microRNAs [39, 40]. The circCD44 was demonstrated to promote tumorigenesis via sponging miR-502-5p in triple-negative breast cancer [39]. Overexpression of circLRP6 accelerated the development of atherosclerosis by sponging miR-145 [40].
Protein interactions
CircRNAs can directly interact with circRNA binding proteins (cRBPs). By binding with proteins, the circRNAs can regulate the translocation of certain proteins. For example, circFOXO3 interacted with stress and senescence associated proteins, preventing the nuclear translocation of these proteins and suppressing their roles against senescence and stress in cardiac fibroblasts [11]. In addition, circRNAs can enhance or dissociate the interactions between two proteins as a facilitator or competitor. The circYAP enhances the interaction between tropomyosin-4 and γ-actin, leading to decrease in actin polymerization and the consequent cardiac fibrosis [32]. The circCCNB1 interacts with CCNB1 and CDK1 proteins and dissociates CCNB1/CDK1 complex, resulting in loss-of-function of CCNB1 on tumorigenesis [41]. All these findings show that circRNAs can regulate gene functions by interacting with proteins.
Forming regulatory complex
CircRNAs also form complexes with mRNA or chromatin upon the circRNA-protein complex. A circRNA may form a ternary complex with protein and mRNA to regulate mRNA stability or protein translation. The formation of circNSUN2-IGF2BP2 protein-HMGA2 RNA complex promotes the interaction of IGF2BP2 and HMGA2, thus enhances the stability of HMGA2 mRNA, and facilitates liver metastasis of colorectal cancer [15]. The circYAP can form a complex with YAP mRNA and two translation initiation machinery proteins, PABP and eIF4G, through which it blocks translation initiation of YAP protein [42]. A circRNA can also form a complex with protein and chromatin to regulate gene transcription. The circZNF827 engaged in a complex containing hnRNP-ZNF827 and chromatin to block the transcription of nerve growth factor receptors during neuronal differentiation [43]. The circACTN4 can recruit YBX1 to FZD7 promoter forming circRNA-protein-chromatin complexes to co-activate FZD7 transcription [44].
Transcriptional regulation
Some ecircRNAs, such as circZNF827 and circACTN4, which are located in the nucleus, can activate gene transcription by interacting with the promoter and recruiting transcription regulator proteins to the promoter region [43, 44]. In addition, ciRNAs such as ci-ankrd52, and EIciRNAs, are found to regulate transcription by binding to RNA polymerase II in the nucleus of human cells [16, 45]. Moreover, some circRNAs functionally regulate gene expression by competing with linear splicing. For instance, during circMBl formation, circMBl competes with pre-MBL mRNA splicing, and adversely affects canonical splicing [6].
Translation
CircRNAs can be translated in a cap-independent manner [46]. Two cap-independent manners have been identified to mediate the translation of circRNAs, which are internal ribosome entry site (IRES) mediated and N6-methyladenosine mediated. IRESs are sequences that recruit initiation factors or ribosomes to mRNA for cap-independent translation [47]. The circZNF609 contains a 753nt ORF for translation to encode a protein via IRES-mediated cap independence [48]. The circMBL3 can encode a protein of 37 kDa via IRES-dependent translation [49]. Some circRNAs are translated independent of IRES but rely on the N6-methyladenosine [50]. The proteins translated from circRNAs play important roles in different organs and diseases. For example, the peptide FBXW7-185aa originating from circ-FBXW7 inhibits cell cycle progression [51]. Work from our lab demonstrated that circNlgn regulated cardiac remodeling by producing a new protein isoform with a smaller size and 9 additional amino acids being translated beyond the splicing site [31].
Functions of circRNAs
The tissue- and age-specific expression of circRNAs suggests their potential roles in tissue development and cell differentiation [52, 53]. Many studies have shown that knockdown or overexpression of circRNAs regulate cell proliferation independently of their cognate linear RNAs [5, 11, 54,55,56,57]. The depletion of circHIPK3 suppresses cell proliferation by sponging multiple microRNAs in HEK293T cells [54]. The circFOXO3 is able to interact with CDK2 and p21, and this triple complex inhibits cell proliferation by increasing the inhibitory effect of p21 on CDK2 [55]. There is evidence that circRNAs play a direct role in immune regulation. The circPAN3 regulates the regeneration of human and mouse LGR5+ intestinal stem cells and activates the WNT signaling pathway [58]. Different circRNAs can regulate cellular stress positively or negatively [59]. The circANRIL has been shown to trigger prolonged nucleolar stress and is associated with an increased risk of atherosclerosis [60]. The highly enriched circRNA expression may be due to the stability of circRNA and the slow division of these cells [5]. Disruption of circRNAs has been reported in neurodegenerative diseases, such as Alzheimer’s disease and autism spectrum disorder [61, 62]. Taken together, most circRNAs specifically express in particular tissues or cells, or at different stages of development for their distinct physiological functions. The physiological and pathological functions of circRNAs are summarized in Fig. 2.
CircRNAs have been reported to play important roles in regulating tissue development, cell proliferation, innate immunity, autophagy, and neuronal functions under physiological conditions. CircRNAs are also involved in the onset and progression of diseases, including cancers, cardiovascular diseases, metabolic diseases, inflammation, and neurodegenerative diseases.
Tissue- and developmental stage-specific expression of circRNAs
Deep sequencing has revealed the characteristics of thousands of circRNAs from a range of tissues and organisms [63]. CircRNAs have been reported to be highly cell type and tissue-specific [53]. A comprehensive classification and characterization of circRNAs in specific tissues or at developmental stages help significantly for their functional studies [53, 64]. CircRNA expression has been investigated in various human adult and fetal tissues. The abundance and expression levels of circRNAs in fetal tissues are relatively higher than in adult tissues, suggesting that circRNAs may function in a tissue-specific and development-specific manner [65].
Tissue-Specific CircRNA Database (TSCD) (http://gb.whu.edu.cn/TSCD) is a comprehensive dataset for tissue-specific expression of circRNAs [53]. The highly abundant tissue-specific circRNAs in human adult, human fetus, and the mouse, obtained from TSCD, are listed in Table 1. Tissue-specific circRNAs are thought to be largely related to tissue development and differentiation. Some circRNAs are expressed in all tissues, while highly abundant circRNAs are expressed differently in various organs [65]. According to TSCD, circRNAs are expressed most highly and specifically in the human brain, and secondly in liver and heart compared to other tissues. In general, there is a significant enrichment in clusters related to the biological functions of host genes of organ-specific circRNAs in related tissues [66]. For example, brain circRNAs are enriched in the region of synaptic transmission [67] and related to the process of aging [65], developmental and adult neurogenesis [68], neuropsychiatric disorders [69], and brain tumors [70]. It has also been reported that some circRNAs are enriched in the heart for cardiac remodelling [31], stress response [11], and endothelium-mesenchyme transition [71]. Other examples of organ-specific circRNA enrichment involve in urea cycle in the liver, striated muscle myosin thick filament, and lung fibrosis [66, 72]. The circRNAs that are expressed in different tissues are summarized in Fig. 3. Some circRNAs are also expressed at different stages of human cancers [73, 74]. A cancer-specific circRNA database (CSCD, http://gb.whu.edu.cn/CSCD) contains different examples, especially cancer-specific circRNAs [75]. Understanding the potential functions of tissue-specific circRNAs is essential for the diagnosis and treatment of diseases. Here we discuss several most studied, highly abundant circRNAs for their tissue-specific and stage-specific expression in diseases. The association of their specific expression, biological functions, and functional mechanisms are summarised in Fig. 4.
The most studied, highly abundant circRNAs are demonstrated to be expressed tissue-specifically or developmental stage-specifically. Their expression in different organs, functions, and mechanisms are listed. Red lines: microRNA sponge. Blue lines: protein interaction. Green line: transcriptional regulation. Yellow line: translation.
ciRS-7/CDR1ase
CDR1as is the best characterized circRNA and fairly abundant in neurons [8] with specificity for complex tissue, cell type, and developmental stage [76]. CDR1as expression was the highest in brain tissue and much less in other tissues under physiological conditions [67]. CDR1as expression in the midbrain suggests its relationship with neurodegenerative diseases [77]. Recent research has revealed that CDR1as has crucial roles in brain development [78] and Alzheimer’s disease (AD) [79]. Other than in brain, studies also reported functions of CDR1as in other diseases including liver cancer and myocardial infarction [80, 81]. However, due to the high abundance of CDR1as/ciRS-7 in the brain, the important physiological and pathological functions of CDR1as/ciRS-7 are identified in brain development and neurological disorders, which implies a link between the tissue-specific expression of CDR1as/ciRS-7 and its functions.
circFOXO3
The circFOXO3 is encoded by the transcription factor FOXO3 gene, which is mainly localized in the cytoplasm and serves as a scaffold for binding to various cRBPs to form stable complexes [55]. The circFOXO3 is highly expressed in the tissues of aged mice that promotes cellular aging in vitro and decreases cardiac functions in vivo via interaction with ID1, E2F1, FAK, and HIF1α [11]. The circFOXO3 is associated with cell cycle progression proliferation [20, 55]. In peripheral blood, circFOXO3 was found to specifically express in aged people compared to younger ones [82]. It has also been suggested that circFOXO3 plays a role in the differentiation of C2C12 myoblast cells, and may provide new clues for the molecular basis of skeletal muscle development [83]. All these studies suggest that circFOXO3 may have stage-specific functions that associated with human development and senescence.
circHIPK3
The circHIPK3 has been reported to be preferably localized in the cytoplasm and is stably and abundantly expressed in various human tissues [54]. It has been stated that circHIPK3 is mostly expressed in the brain, especially in the cerebellum [84], and is one of the most abundant circulating RNAs in pancreatic islets [85]. In addition, circHIPK3 is one of the highly expressed and conserved circRNAs in the heart and is associated with cell proliferation, migration, angiogenesis in the pathogenesis of various cardiovascular diseases [86]. The circHIPK3 is also the most abundant circRNAs in human colon, lung, and stomach tissues [65]. CircHIPK3 may be involved in intestinal epithelium homeostasis, fibroblast-myofibroblast transition, and tumorigenesis [54, 87, 88].
circCDYL
The expression of circCDYL, a circular RNA originated from back insertion from the fourth exon of the CDYL gene, was found to be decreased in myocardial tissues and hypoxia myocardial cells [89]. It is a new regulator of myocardial formation, and it improves cardiac function during myocardial regeneration after acute myocardial infarction [89]. The circCDYL was found to be highly expressed in different types of cancers, especially in the early stages of hepatocellular carcinoma [73], bladder cancers [74], and breast cancer [90]. Upregulation of circCDYL in the serum and tumor tissue of cancer patients suggested circCDYL might be a potential biomarker for an early stage of cancers.
Other circRNAs
To better understand the tissue-specific expression of circRNAs, some studies conducted a landscape profiling of circRNAs in specific organs. Tan et al. analyzed the circRNA profile in human and mouse hearts. 1664 cardiac-specific circRNAs were identified in human heart [91]. These cardiac-specific circRNAs were confirmed to be actively involved in a variety of heart-specific physiological processes. Some of the highest cardiac expressed circRNAs are also highly enriched in liver, muscles, and brain [91], while some are specific to the fetal heart [92]. Some other studies also found the expression rates of circZkscan1 in liver are much higher than other tissues [65]. These findings suggest the tissue-specific pattern of circRNA may be associated with their special functions in these tissues. The tissue-specific expression and stability offer important functions for these molecules. The expression of various circRNAs in different organs and stages are shown in Table 2. Although we do not yet understand all the mechanisms responsible for regulating cell-type-specific isoform expression and diversity, it may not be surprising to detect different isoforms of circRNAs when alternative splicing occurs and thousands of isoforms can be produced from a single gene.
Potential reasons for circRNA abundancy
Neuronal development
CircRNAs exhibit different expression levels during development. Studies in Drosophila showed that the nervous tissues are highly enriched for circRNAs [1]. In rat, only 72 circRNAs were expressed in all organs throughout the whole development stage [93]. Another study reported that circRNAs might be differentially regulated during development in the mouse hippocampus [94]. Slow cell-turnover and limited regeneration are thought to contribute to the stability of circRNAs. Moreover, some circRNAs are abundant at synapses, and synaptic neuropil revealed increased expression of circRNAs [8, 94]. This may be one of the reasons why higher number of circRNAs are observed in humans compared to mouse brains, as the synaptic density in the human cerebral cortex is higher than that in the mouse brain [95]. Another possible reason may include that neuronal genes have long introns and expression of circRNAs can be conserved by long introns [96]. Transcription rate, RNA turnover, and cell division are influenced by circRNA expression in neuron tissue [97]. High levels of circRNA expression are observed in the cortex in early and mid-pregnancy when neurogenesis occurs [98].
Culturing conditions and stress regulate circRNA abundance
Accumulating evidence has demonstrated that cellular conditions and stress might regulate circRNA expression. The proliferative status of cells is an effective factor on circRNA expression. Proliferating cells, including cancer cells, express lower levels of circRNA [72]. Song et al. revealed that human glioma tissues have less amounts of circRNAs than healthy brain tissues, which may result from the collapse of the back-splicing events under abnormal conditions [99]. CircRNAs can be regulated by stress events such as at high and low temperatures, or under oxidative stress [6, 11]. It has been reported that under stress, circRNA levels could potentially be altered by factors that bind and modulate intronic sequences [59]. For instance, Qiu et al. reported that ADAR1 modulates circRNA expression during endoplasmic reticulum stress [100]. However, the mechanisms regulating circRNA levels during stress have not been fully understood.
Aging
RNA-seq studies have shown that circRNA levels are greatly increased during aging, and multiple mechanisms are involved in the accumulation of circRNAs associated with aging tissues [101]. Firstly, circRNAs are resistant to degradation from exoribonucleases and have high stability [96], leading to accumulation. Secondly, circRNA biogenesis might be affected by age-related changes in alternative splicing. CircRNAs are produced by alternative splicing, and thus it is reasonable to assume that age-related changes in back-splicing, particularly splice factors, affect alternative splicing results. For example, circRNA expression is regulated by the FUS insertion factor in cultured mouse motor neurons [102]. In addition, other mechanisms involved in circRNA biogenesis may play a role during aging, such as RBPs, which influence the stability of circRNAs [101]. Harries et al. demonstrated that SRFR6 and SRFS1 splice factors were downregulated during aging, which may be associated with the accumulation of circRNAs [103]. Zhou et al. showed that circRNAs may be necessary to regulate the spermatogenesis process in testicular tissue, which is the most sensitive organ showing age-related changes, and age-related changes in circRNA levels [104].
Potential reasons for tissue-specific expression of circRNAs
CircRNAs are conserved across species and often exhibit cell type- or tissue-specific expression. However, the pattern of differential expression of circRNAs across species and the underlying mechanism is still under investigation. The following factors may be considered when the tissue-specific expression of circRNAs is discussed.
Host gene expression
CircRNAs are generally known to be specific to cells and tissues, but in some cases, a circRNA is reported to have a function in a particular tissue while the host gene has important physiological roles in a different tissue [53]. Gene ontology analysis of host genes was performed in different organs to understand the possible functions of organ-specific circRNAs [93]. For example, the brain host gene of a circRNA would be involved in neuronal secretion and synaptic functions, while cardiac muscle differentiation and development in the heart also activate the production of certain circRNAs that may play a role in muscle contraction. From this point of view, it is necessary to investigate whether the host gene has a function in the tissue as well.
Organ-specific functions
Various studies have shown that circRNAs are highly expressed in neuron/brain tissues and tissue-specific [8, 94]. Moreover, the abundance appears to be dependent on the biogenesis and functions of circRNA. Many circRNAs in the brain have been reported to play a role in neurotransmitter functions, neuron maturation, and synaptic activity [93]. For example, circHomer 1 originating from the Homer Scaffolding Protein 1 pre-mRNA is demonstrated to regulate neuronal plasticity and structural changes in the synapse during development [94]. Xia et al. reported that the gene encoding the protein Corin produced 40 heart-specific circRNA isoforms. Among them, 25 isoforms are specific to adults, suggesting that circRNAs from this gene are specific to the functions of the adult heart. Albumin, as another example, generates 160 liver-specific circRNA isoforms, 95 of which are specific for the adult liver and 33 are specific for the fetal liver. This suggests that tissue-specific circRNAs may be related to the functions in development and differentiation [53].
Specific supporting systems in different organs
The feature of highly tissue-specific expression of circRNA raises the question: What determines this tissue-preferred expression pattern? How is the specific expression of circRNAs regulated? CircRNA biogenesis is precisely controlled by the presence of many cis-acting elements and trans-acting factors to maintain physiological balance within cell [105]. Repetitive elements (cis-elements), especially human Alu sequences, play roles in circRNA biogenesis [106]. The back-splicing exons are associated with Alu elements in the surrounding long introns [96]. Alu has the potential to create inverted repetitive Alu sequences [107], and IRAlus supports circRNA production leading to alternative cyclization [106]. The repetitive elements are responsible for most circRNA formation in humans [108].
Apart from cis-elements, various protein factors positively or negatively regulate circRNA biogenesis [6, 8]. It is stated that RBPs are key regulators of the tissue-preferred expression pattern of circRNAs [109]. The first RBP identified to regulate circRNA expression was Muscleblind protein in Drosophila [6]. MBL plays a role in various RNA processing steps, including mammalian homologues, RNA localization, alternative polyadenylation, and alternative splicing [110]. Immune factors NF90/NF110, each containing two dsRNA binding domains, can directly bind to the IRAlus formed in pre-mRNA, promoting circRNA formation [36]. Expression of double-stranded RNA-specific adenosine deaminase-1 (ADAR1) destabilizes the base-pairing interactions required for circRNA biogenesis and reduces circRNA expression [8]. A nuclear RNA helicase-9 (DHX9) containing both a dsRBD and an RNA helicase domain has been shown to inhibit circRNA production by unraveling RNA structures and by binding directly to Alu elements to regulate circRNA-producing genes, RNA processing, and translation [111]. Many circRNAs are also generated through splicing factor Quaking, which lack dsRBD during human epithelial-mesenchymal transition [36, 112]. Regarding tissue-specific regulation, it has been reported that heterogeneous nuclear ribonucleoprotein-L plays roles in circRNA regulation in human prostate cancer, and RNA-binding motif protein-20 (RBM20) is essential for the formation of a subset circRNA that originate from the Titin gene in mammalian hearts [113, 114]. It reveals that circRNA biogenesis is highly dependent on the biological setting and is tightly regulated in tissues and cells using different cis-elements and trans factors.
circRNA-based biomarkers and therapeutic strategies
The ability to characterize circRNAs function in a tissue-specific manner is crucial to studying disease mechanisms. Due to the high stability and tissue- or cell-type-specific expression, circRNAs have emerged as promising biomarkers and therapeutic targets [115].
CircRNAs serve as prognosis related biomarkers
The circRNAs have much longer half-life than linear RNAs [116], which make them ideal candidates for being biomarkers. Studies have revealed the potential of circRNAs serving as diagnostic biomarkers in various diseases [117]. More than 2400 circRNAs have been detected in human whole blood. In addition, the total circRNA levels in human blood are comparable to those in the brain, indicating the high abundance of circRNAs in blood and the great potential for using circRNAs as biomarkers [118]. Researchers are seeking the application of circRNAs as biomarkers for early stage diagnosis or prognosis. In patients with acute myocardial infarction, circRNA levels were identified to be significantly lower than healthy controls in peripheral blood and strongly predicted the dysfunction of left ventricle in these patients [119]. In addition, a significant decrease in circ_0003391 was found in the peripheral blood of AD patients, suggesting circ_0003391 might be used to distinguish AD patients from other dementia [120]. Recently, an atlas of blood-based biomarkers for early diagnosis of cancers (http://bbcancer.renlab.org/) has been established. It includes expression data of different types of RNAs across different types of cancers from 7184 samples, among which 5040 are blood samples [121]. Other than serving as biomarkers for diagnosis, circRNAs can also be used for predicting the prognosis. The circ-KIAA1244 was identified to have lower expression levels in the plasma of gastric cancer patients and negatively correlated with metastasis stage and patient lifespan [122]. Another study in acute ischemic stroke found levels of circRNAs were significantly increased in patients’ plasma and the outcomes can be predicted by the changes of these circRNAs [123].
Exosome derived circRNAs
Exosomes are extracellular vehicles, widely presenting in blood and other body fluids, that carry a variety of nucleic acid molecules, proteins, and signaling lipids to assist the intercellular communications [124, 125]. The circRNAs are found to be enriched in exosomes, and a large number of circRNAs were identified in human serum exosomes [125,126,127]. The expression profiles of exosomal circRNAs in colon cancer could distinguish patients from the healthy individuals, suggesting the potential of exosomal circRNAs serve as biomarkers in cancer diagnosis [127]. The serum exosomal circIARS levels were significantly increased in the patients with metastatic pancreatic cancer and also correlated with the TNM stages and poor prognosis, which suggested serum exosomal circIARS could be a potential non-invasive biomarker [128]. Exosomal circRNA can also be used for early diagnosis of immune diseases. The levels of exosomal hsa_circ_0087862 and hsa_circ_0012077 in cerebrospinal fluid were identified to be biomarkers for immune-mediated demyelinating disease [129]. Several databases are employed for searching circRNAs in human body fluid. A database exoRBase was created showing the abundance and diversity of circRNAs in exosomes of human biofluid [130, 131].
Using circRNA as therapeutic strategies
Due to the tissue-specific and cell type-specific expression, different circRNAs have been identified to serve as therapeutic targets in various diseases including cardiovascular diseases, nervous system disorders, cancers, musculoskeletal diseases, etc [117]. With the explosive growth and successful application of RNA therapy in the recent years, researchers are eager to seek better RNA candidates which are more stable and less immune-stimulatory than mRNA. CircRNAs are more resistant to exonucleases than linear RNAs, and lack of free-end allows endogenous circRNAs to escape from RNA-mediated innate immune response [132]. Although there are controversial opinions in the immunogenicity of circRNA, it is still regarded as one of the most promising candidates for RNA therapy.
For effectively and safely delivering the circRNAs to the target organs, various techniques have been employed including adeno-associated virus (AAV), nanoparticles, adenovirus, and plasmids [133,134,135]. The circRNAs can be delivered to multiple tissues in vivo using recombinant AAV vectors, thereby providing a proof-of-concept study for in vivo delivery of artificial or natural circRNA [134]. Our team successfully deliver several nanoparticle-conjugated circRNA plasmids and siRNAs into mice [31, 32]. As one of the most challenging aspects of circRNA therapy, delivering circRNA to specific organs or tissues for precise treatment is of great importance.
Methods to overexpress or knockdown circRNAs
Researchers have developed different methods to either overexpress or knockdown circRNAs in vitro and in vivo [18, 136]. Overexpression can be performed by delivering plasmids containing circRNA-producing exons in animals or cells [31, 32]. Synthetic circRNAs that are transcribed and cyclized in vitro and delivered to specific tissue with AAV is another therapeutic approach. Synthetic circRNAs have been engineered to contain more microRNA binding sites or pursue long-term and large-scale translation [137, 138]. Compared to gain-of-function studies with plasmids, researchers encounter much more challenges in circRNA knockdown due to the shared sequences between circRNA and its linear cognate. Small interfering RNAs (siRNA), short hairpin RNAs (shRNA), CRISPR/Cas9, and RNA-targeting Cas13 system have been successfully used to silence tissue-specific circRNAs in mice [67, 117, 139]. The shRNAs and siRNAs have been widely used in a large body of circRNA functional studies to trigger RNA interference for a variety of circRNAs in vitro and in vivo [117]. CRISPR/Cas9 system has also been used to knockdown circRNAs in different cells and establishes the first circRNA knockout mice model of CDR1as without apparent off-target effect [67]. CRISPR/Cas13 was reported to effectively knockdown circRNAs expression without affecting their cognate mRNA by the guide RNAs targeting back-splicing junction [139]. The therapeutic strategies of circRNAs may contribute to tissue-specific treatment.
Conclusion and perspective
CircRNAs have been acknowledged to be stable, abundant, conserved, and often exhibit tissue-specific and developmental stage-specific expression. Based on these advantages, the focuses in this field are not limited in functional and mechanistic studies but also expanded to the applications of circRNAs in pre-clinical and clinical therapy of human diseases. Advanced RNA-seq and bioinformatics technologies have exposed the prevalence of circRNAs in eukaryotic transcriptomes, and these molecules have become an important research hotspot.
Although progress has been made in identifying and characterizing circRNAs, many more questions have arisen than answers. Firstly, circRNA biogenesis is elaborately processed by cis-elements and trans-acting factors in normal cells, and the change of the state of these factors in pathological conditions can accelerate or suppress circRNA production. However, our understanding of the function-related circRNA biogenesis and localization, and specifically how tissue-specific expression contributes to their unique regulatory patterns in various biological processes are limited. What regulates the tissue-specific expression of circRNAs and the association between the specificity and abundance in certain tissues and stages remain ambiguous.
It’s of great interest to investigate how the findings from circRNA studies can be applied in the clinical scenarios for early diagnosis and predicting prognosis of certain human diseases. Currently, many studies have identified the levels of specific circRNAs are elevated or downregulated in certain types of body fluid or circulating extracellular vesicles and have corroborated the possible correlation of such circRNAs with certain diseases. Nevertheless, the data derived from a large scale of clinical samples are still needed to validate the reliability and sensitivity of these circRNA biomarkers.
The tissue-specific expression pattern of circRNA could be sine qua non for delivery of circRNAs to specific organs or cell types in gene therapies, since it is a precise and cost-effective way for circRNA therapy. The current delivery methods in pre-clinical circRNA therapy are limited to systemic delivery via intravenous or intraperitoneal injection or administering the circRNAs direct into targeting organs. For the aim of therapeutic application in terms of specificity and efficacy, new methods based on the feature of tissue-specific expression of individual circRNA are expected to be explored in the future via systemic administration but targeting to certain organs in a specific disease. Taken together, further understanding and profiling the tissue-specific and stage-specific expression patterns of circRNAs may lay the ground for the applications of circRNAs in the therapy of human diseases.
References
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–80.
Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PloS One. 2014;9:e90859.
Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109:401–7.
Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–9.
Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777.
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85.
Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61.
Du WW, Yang W, Chen Y, Wu Z-K, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–12.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503–17.
Zhang J, Zhang X, Li C, Yue L, Ding N, Riordan T, et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019;16:220–32.
Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–44.
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Wang S, Latallo MJ, Zhang Z, Huang B, Bobrovnikov DG, Dong D, et al. Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD. Nat Commun. 2021;12:4908.
Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16:503–14.
Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: An emerging key player in RNA world. Brief Bioinforma. 2017;18:547–57.
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–70.
Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409.
Pandey PR, Munk R, Kundu G, De S, Abdelmohsen K, Gorospe M. Methods for analysis of circular RNAs. Wiley Interdiscip Rev RNA. 2020;11:e1566.
Hansen TB, Venø MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58.
Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985.
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Liu Y-C, Li J-R, Sun C-H, Andrews E, Chao R-F, Lin F-M, et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–15.
Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905.
Schneider T, Schreiner S, Preusser C, Bindereif A, Rossbach O. Northern blot analysis of circular RNAs. Methods Mol Biol. 2018;1724:119–33.
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85–96.
Du WW, Xu J, Yang W, Wu N, Li F, Zhou L, et al. A neuroligin isoform translated by circnlgn contributes to cardiac remodeling. Circ Res. 2021;129:568–82.
Wu N, Xu J, Du WW, Li X, Awan FM, Li F, et al. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with Tropomyosin-4 and Gamma-Actin Decreasing Actin Polymerization. Mol Ther. 2021;29:1138–50.
Du WW, Yang W, Li X, Fang L, Wu N, Li F, et al. The circular RNA circSKA3 binds Integrin beta1 to induce invadopodium formation enhancing breast cancer invasion. Mol Ther: J Am Soc Gene Ther. 2020;28:1287–98.
Fang L, Du WW, Lyu J, Dong J, Zhang C, Yang W, et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell death Differ. 2018;25:2195–208.
Li F, Yang Q, He AT, Yang BB. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin Cancer Biol. 2021;75:49–61.
Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–42.
Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–31.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B, et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. 2021;20:138.
Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circulation Res. 2019;124:498–510.
Fang L, Du WW, Awan FM, Dong J, Yang BB. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–26.
Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang C, et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758–73.
Hollensen AK, Thomsen HS, Lloret-Llinares M, Kamstrup AB, Jensen JM, Luckmann M, et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. Elife. 2020;9:e58478.
Chen Q, Wang H, Li Z, Li F, Liang L, Zou Y, et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol. 2022;76:135–47.
Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806.
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475.
Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–5.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9.
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol cell. 2017;66:9–21.e7.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–15.
Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–91.
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinforma. 2017;18:984–92.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–20.
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, et al. The Circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther: J Am Soc Gene Ther. 2017;25:2062–74.
Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20:183–94.
Fischer JW, Leung AKL. CircRNAs: A regulator of cellular stress. Crit Rev Biochem Mol Biol. 2017;52:220–33.
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903–12.
Chen Y-J, Chen C-Y, Mai T-L, Chuang C-F, Chen Y-C, Gupta SK, et al. Genome-wide, integrative analysis of circular RNA dysregulation and the corresponding circular RNA-microRNA-mRNA regulatory axes in autism. Genome Res. 2020;30:375–91.
Werfel S, Nothjunge S, Schwarzmayr T, Strom T-M, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–7.
Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J. et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444–60.
Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: A study using RNA-seq data. BMC Genomics. 2017;18:680.
Mahmoudi E, Cairns MJ. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci Rep. 2019;9:2564.
Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science (New York, N.Y.). 2017;357:eaam8526.
Chen BJ, Huang S, Janitz M. Changes in circular RNA expression patterns during human foetal brain development. Genomics. 2019;111:753–8.
Reddy AS, O’Brien D, Pisat N, Weichselbaum CT, Sakers K, Lisci M, et al. A comprehensive analysis of cell type-specific nuclear RNA from neurons and glia of the brain. Biol Psychiatry. 2017;81:252–64.
Xu K, Ding L, Chang T-C, Shao Y, Chiang J, Mulder H, et al. Structure and evolution of double minutes in diagnosis and relapse brain tumors. Acta Neuropathologica. 2019;137:123–37.
Hulshoff MS, Xu X, Krenning G, Zeisberg EM. Epigenetic regulation of endothelial-to-mesenchymal transition in chronic heart disease. Arteriosclerosis, Thrombosis, Vasc Biol. 2018;38:1986–96.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory rnas network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatol (Baltim, Md). 2020;71:130–47.
Okholm TLH, Nielsen MM, Hamilton MP, Christensen L-L, Vang S, Hedegaard J, et al. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2017;2:36.
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, et al. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929.
Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep. 2015;33:2669–74.
Azari H, Mousavi P, Karimi E, Sadri F, Zarei M, Rafat M, et al. The expanding role of CDR1-AS in the regulation and development of cancer and human diseases. J Cell Physiol. 2021;236:771–90.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307.
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11:8183–203.
Geng H-H, Li R, Su Y-M, Xiao J, Pan M, Cai X-X, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PloS One. 2016;11:e0151753.
Haque S, Ames RM, Moore K, Pilling LC, Peters LL, Bandinelli S, et al. circRNAs expressed in human peripheral blood are associated with human aging phenotypes, cellular senescence and mouse lifespan. Geroscience. 2020;42:183–99.
Li X, Li C, Liu Z, Ni W, Yao R, Xu Y, et al. Circular RNA circ-FoxO3 inhibits myoblast cells differentiation. Cells. 2019;8:616.
Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–47.
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Venø MT, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab. 2018;9:69–83.
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. 2019;292:188–96.
Xiao L, Ma XX, Luo J, Chung HK, Kwon MS, Yu TX, et al. Circular RNA CircHIPK3 promotes homeostasis of the intestinal epithelium by reducing MicroRNA 29b function. Gastroenterology. 2021;161:1303–17 e3.
Zhang J-X, Lu J, Xie H, Wang D-P, Ni H-E, Zhu Y, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10:182.
Zhang M, Wang Z, Cheng Q, Wang Z, Lv X, Wang Z, et al. Circular RNA (circRNA) CDYL induces myocardial regeneration by ceRNA after myocardial infarction. Med Sci Monit: Int Med J Exp Clin Res. 2020;26:e923188.
Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65.
Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309.
Lee ECS, Elhassan SAM, Lim GPL, Kok WH, Tan SW, Leong EN, et al. The roles of circular RNAs in human development and diseases. Biomed Pharmacother. 2019;111:198–208.
Tang M, Kui L, Lu G, Chen W. Disease-associated circular RNAs: From biology to computational identification. BioMed Res Int. 2020;2020:6798590.
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–10.
Herculano-Houzel S. The human brain in numbers: A linearly scaled-up primate brain. Front Hum Neurosci. 2009;3:31.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (N. Y, N. Y). 2013;19:141–57.
Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PloS One. 2010;5:e11173.
Di Timoteo G, Rossi F, Bozzoni I. Circular RNAs in cell differentiation and development. Development. 2020;147:dev182725.
Song X, Zhang N, Han P, Moon B-S, Lai RK, Wang K, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87.
Qiu W, Wang X, Buchanan M, He K, Sharma R, Zhang L, et al. ADAR1 is essential for intestinal homeostasis and stem cell maintenance. Cell Death Dis. 2013;4:e599.
Knupp D, Miura P. CircRNA accumulation: A new hallmark of aging? Mechanisms Ageing Dev. 2018;173:71–79.
Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741.
Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10:868–78.
Zhou T, Xie X, Li M, Shi J, Zhou JJ, Knox KS, et al. Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA (N. Y, N. Y). 2018;24:1443–56.
Wawrzyniak O, Zarębska Ż, Kuczyński K, Gotz-Więckowska A, Rolle K. Protein-related circular RNAs in human pathologies. Cells. 2020;9:1841.
Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47.
Chen L-L, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–705.
Dong R, Ma X-K, Chen L-L, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–74.
Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: Old players and new actors. Trends Cancer. 2017;3:506–28.
Cortés-López M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–37.
Wu Q, Li P, Wu M, Liu Q. Deregulation of circular RNAs in cancer from the perspectives of aberrant biogenesis, transport and removal. Front Genet. 2019;10:16.
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34.
Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA. 2017;114:E5207–E5215.
Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, et al. RBM20 regulates circular RNA production from the Titin gene. Circ Res. 2016;119:996–1003.
He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct Target Ther. 2021;6:185.
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83.
Yang Q, Li F, He AT, Yang BB. Circular RNAs: Expression, localization, and therapeutic potentials. Mol Ther. 2021;29:1683–702.
Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214.
Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247–8.
Liu L, Chen X, Chen YH, Zhang K. Identification of circular RNA hsa_Circ_0003391 in peripheral blood is potentially associated with Alzheimer’s disease. Front Aging Neurosci. 2020;12:601965.
Zuo Z, Hu H, Xu Q, Luo X, Peng D, Zhu K, et al. BBCancer: an expression atlas of blood-based biomarkers in the early diagnosis of cancers. Nucleic Acids Res. 2020;48:D789–D96.
Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137.
Zuo L, Zhang L, Zu J, Wang Z, Han B, Chen B, et al. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke. 2020;51:319–23.
van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28.
Du WW, Li X, Ma J, Fang L, Wu N, Li F, et al. Promotion of tumor progression by exosome transmission of circular RNA circSKA3. Mol Ther Nucleic Acids. 2022;27:276–92.
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4.
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177.
He J, Ren M, Li H, Yang L, Wang X, Yang Q. Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front Genet. 2019;10:860.
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112.
Lai H, Li Y, Zhang H, Hu J, Liao J, Su Y, et al. exoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022;50:D118–D128.
Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang Y, Bisaria N, Anderson DG. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 2019;74:508–20 e4.
Ma J, Du WW, Zeng K, Wu N, Fang L, Lyu J, et al. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther: J Am Soc Gene Ther. 2021;29:2754–68.
Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF, et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol Ther Nucleic Acids. 2018;13:89–98.
Sun K, Wang D, Yang BB, Ma J. The emerging functions of circular RNAs in bladder cancer. Cancers. 2021;13:4618.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Rossbach O. Artificial circular RNA sponges targeting MicroRNAs as a novel tool in molecular biology. Mol Ther Nucleic Acids. 2019;17:452–4.
Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629.
Li S, Li X, Xue W, Zhang L, Yang LZ, Cao SM, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. 2020;18:51–9.
Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–76.
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–55.
Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907.
Funding
This work was supported by grants from Canadian Institutes of Health Research (PJT-153105, PJT-155962, and PJT-166107) to BBY.
Author information
Authors and Affiliations
Contributions
SM, NW, and BBY designed the structure of the review and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by H.-U. Simon
Rights and permissions
About this article
Cite this article
Misir, S., Wu, N. & Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ 29, 481–491 (2022). https://doi.org/10.1038/s41418-022-00948-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00948-7
This article is cited by
-
Hsa_circ_0004872 alleviates meningioma progression by sponging miR-190a-3p/PTEN signaling
BMC Cancer (2024)
-
Non-coding RNA in the gut of the blood-feeding parasitic worm, Haemonchus contortus
Veterinary Research (2024)
-
Circular RNAs and cervical cancer: friends or foes? A landscape on circRNA-mediated regulation of key signaling pathways involved in the onset and progression of HPV-related cervical neoplasms
Cell Communication and Signaling (2024)
-
Hsa_circ_0007990 promotes breast cancer growth via inhibiting YBX1 protein degradation to activate E2F1 transcription
Cell Death & Disease (2024)
-
Lymphatic vessel: origin, heterogeneity, biological functions, and therapeutic targets
Signal Transduction and Targeted Therapy (2024)