Abstract
The Shwachman-Diamond syndrome (SDS) is characterized by exocrine pancreatic insufficiency, neutrophil defect, and skeletal abnormalities. The molecular basis for this syndrome was recently identified as a defect in a novel nucleolar protein termed the Shwachman-Bodian-Diamond syndrome (SBDS) protein. Beyond human pathologic descriptions, there are little data addressing the role of SBDS during pancreas and granulocytes development. We hypothesize that sbds gene function is essential for pancreas and myeloid development in the zebrafish. By homology searching, we identified the zebrafish sbds ortholog and then analyzed its expression by reverse transcriptase-polymerase chain reaction and in situ hybridization. We found that the sbds gene is expressed dynamically during development. To study the function of sbds during development, we induced loss of gene function by morpholino-mediated gene knockdown. The knockdown induced a morphogenetic defect in the pancreas, altering the spatial relationship between exocrine and endocrine components. We also noted granulopoiesis defect using myeloperoxidase as a marker. We conclude that sbds function is essential for normal pancreas and myeloid development in zebrafish. These data provide novel insight into the role of the sbds gene and support using zebrafish as a model system to study sbds gene function and for evaluation of novel therapies.
Similar content being viewed by others
Main
Shwachman-Diamond syndrome (SDS) is a rare, multisystem disorder characterized by exocrine pancreatic insufficiency, hematological dysfunction with a high risk for developing acute myeloid leukemia, and skeletal abnormalities (1–5). Additional features that have been reported in some patients includes hepatomegaly with abnormal liver function tests, renal calculi, myocardial fibrosis, behavioral and learning difficulties (6–15). Aside from cystic fibrosis, SDS is the most common cause of inherited exocrine pancreatic insufficiency in children. It is the third most common cause of inherited bone marrow failure. Shwachman syndrome is inherited in an autosomal recessive manner with an incidence of 1 in 76,000 (16). By positional cloning, the Shwachman-Bodian-Diamond syndrome (SBDS) gene was recently identified in the long arm of chromosome 7 near the centromere (17–19). The SBDS gene contains five exons spanning a length of 7.9 kb. SBDS locus contains an adjacent pseudogene (SBDSP) that is nonfunctional, but with 97% nucleotide sequence identity. However, the pseudogene contains deletions and nucleotide changes that are predicted to result in protein truncation. In the majority of patients with SDS, it appears that SBDS and SBDSP recombined resulting in unidirectional gene conversion from the pseudogene to SBDS (17). This gene conversion change is predicted to disrupt the donor splice site of intron 2 and the 8-bp deletion resulting in premature truncation of the encoded protein.
The SBDS gene is predicted to encode a novel 250-amino acid protein. This protein is highly conserved throughout evolution, and orthologs exist in species from plants, yeast, to vertebrate animals (20). Several lines of evidence postulate the role of SBDS protein in RNA metabolism (17,21), including the intranucleolar localization of SBDS protein in human cells (22), but its specific biochemical function remains unknown.
Beyond human pathologic descriptions, there are few data addressing the role of SBDS gene during development of the pancreas or the granulocytes. Impaired pancreatic function in SDS patients has been attributed to abnormal acinar cell development (23). The severity of pancreatic dysfunction is not concordant with blood or bone abnormalities (3). Approximately 50% of SDS patients shows some improvement in pancreatic function with increasing age (23). Imaging and autopsy studies showed the fatty replacement of pancreatic exocrine tissue with preserved ducts and islet architecture (24,25). A recent study found that Sbds is an essential gene in mouse. Mice heterozygous for the gene deletion were overtly normal, whereas Sbds homozygous mutant embryos underwent developmental arrest before implantation. This emphasizes the need to develop an animal model to detect the organ development in a tractable vertebrate (26).
The recent identification of SBDS gene mutation allows us to explore the developmental processes that give rise to the clinical features of this syndrome. We hypothesize that SBDS gene function is essential for the normal pancreas and myeloid development. To test this hypothesis, we used zebrafish as the animal model to study sbds gene function particularly with regard to pancreas and myeloid development. The zebrafish, Danio rerio, offers several distinct advantages as a genetic model system including the high fecundity, external fertilization, short generation time (3–4 mo), easy maintenance, rapid development time, and translucent embryos. Moreover, many zebrafish genes are conserved in human. In this study, we determined the localization of sbds gene expression in the zebrafish and evaluated the effect of gene perturbation on the pancreas and granulocytes. All procedures concerning zebrafish were done in accordance with the protocol that was approved by the Medical College of Wisconsin and Institutional Animal Care and Use Committee.
MATERIALS AND METHODS
Zebrafish embryos.
Wild-type (TUAB strain) zebrafish embryos were obtained from natural spawning of wild-type adults. Embryos were raised, maintained, and staged at 28.5°C according to hours post fertilization (hpf) and morphologic criteria (27).
Morpholino-mediated knockdowns.
We generated morpholino modified antisense oligonucleotides against the sbds gene translation start site, and splice donor sites of exon-intron 1 and 2 (ATG, EI-1, and EI-2, respectively). The splicing interference was detected by reverse transcriptase-polymerase chain reaction (RT-PCR) across the targeted exon. Sequences for the morpholinos and RT-PCR primers are given in Table 1. Morpholinos, at 100 μM, were micro-injected into one-cell stage embryos. Approximately 50 embryos were injected at each sitting, and the observed defect was noted in approximately 70% of the knockdown embryos. All the morpholinos greatly reduced sbds gene expression and produced similar results without nonspecific side effects. Embryos were collected at different hours post fertilization (24, 48, 72, 96, and 120 hpf), dechorionated, fixed in 4% paraformaldehyde (PFA) in PBS for 2 h at room temperature, washed twice in PBS with 0.1% (vol/vol) Tween 20 (PBT), and stored in methanol at −20°C for whole mount in situ hybridization (ISH).
RT-PCR.
Total RNA was extracted from zebrafish embryos using Trizol (Invitrogen). Reverse transcription was performed using synthetic DNA oligo (Poly dT primer). PCR reactions used Taq polymerase from Qiagen, and contained 5% of the reverse transcription.
RNA in situ hybridization.
Probe synthesis and whole-mount RNA in situ hybridization were carried out as described (28). Antisense probes were generated by in vitro transcription using DNA fragments derived from RT-PCR with the T7 start site incorporated into the 5′ tail of the reverse primer (Table 2).
Histologic analysis.
Embryos were fixed in 4% PFA in PBS for 2 h at room temperature or overnight at 4°C. Embryos were then washed in PBS, serially dehydrated in ethanol and then embedded in glycol methacrylate, JB4 (polysciences). Four-micrometer thick sections were stained with hematoxylin and eosin, analyzed using a Zeiss Axioplan microscope, and captured using a Q-imaging digital camera and openLab 4.02 (Improvision).
RESULTS
Sequence and alignment of the zebrafish Sbds ortholog.
We performed a sequence search (tblasn) of the zebrafish genome assembly using the human SBDS protein identified on chromosome 7. We found a gene that is predicted to encode a protein 90% identical to the human orthologs. We found no evidence of a pseudo-gene adjacent to the zebrafish sbds locus. In addition, alignment of the zebrafish and human genomic sequences revealed the same exon-intron boundaries (Fig. 1).
Protein sequence comparison of zebrafish Sbds with other SBDS orthologs. Clustal alignment of zebrafish, human, and yeast sbds orthologs revealed a high degree of homology (90%), with conservation of the intron-exon junctions. These are highlighted by the vertical lines, with flanking numbers denoting exon number. Shaded boxes reflect sequence identity. Accession numbers used for comparison: human, Q3SWZ6; zebrafish, AAH50179; yeast NP 013122.
The sbds gene is highly expressed in zebrafish.
We analyzed the sbds gene expression during zebrafish development at stages 24–120 hpf by RT-PCR and found the gene is expressed throughout these stages as shown in the gel electrophoresis (Fig. 2A).
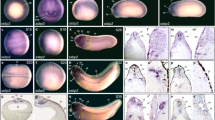
sbds expression analysis (A) RT-PCR analysis of sbds expression at different stages post fertilization (hpf) compared with eIF1α control (B,D,E). Whole-mount in situ hybridizations for sbds in zebrafish embryos at different hours post fertilization. (B) 48 hpf, dorsal view, sbds expression is seen in the branchial arches (BA), liver (L), gut (g), and pancreas (P). (C) 48 hpf, dorsal view of foxA3 and pdx (probe cocktail) shows digestive organ epithelial-specific staining pattern (for comparison with sbds-specific pattern shown in B). (D) 72 hpf expression persists in the gut, liver, and the pancreas. (E) At 120 hpf, the liver is no longer stained, the gut expression is decreased, but pancreatic expression persists. (F–G) Histologic sections of 48 hpf embryos stained for sbds expression and counterstained with DAPI to reveal cell nuclei. (F) Low power view of DAPI-stained section, with box indicating expanded views seen in panels G and H. (G) Transmission light photomicrograph of sbds-specific staining, revealing most of the gene expression localized to cells ventral to the digestive organs. Dotted lines indicate positions of gut tube (g) and pancreas (P). (H) Merged view of DAPI and sbds-stained sections. Scale bars: B,C, 167 μm; D,E, 250 μm; F, 50 μm; G,H, 16 μm.
sbds gene localization.
As an initial step to examine the role of sbds during development, we determined its expression pattern by whole mount in situ hybridization. At 48 hpf, we noticed that the gene is expressed preferentially in the branchial arches and digestive tract, including the liver, pancreas, and gut (Fig. 2B). We compared this staining pattern to markers known to be expressed in the epithelium of these organs, namely foxA3 (endoderm) and pdx (pancreatic bud) (28) (Fig. 2C). We noted an overall similar anatomic pattern of expression between sbds and these genes, but with a notable difference. The expression pattern of sbds was broader and more diffuse than the epithelial markers. To characterize this further, we performed histologic sectioning of the sbds-stained embryos. We found that sbds expression is most prominent in the cells intervening between the organ parenchyma and the yolk (Fig. 2F–H). This location corresponds to the visceral mesenchyme. As development proceeds, sbds expression persists in and around the pancreas, liver, and gut (Fig. 2D). Then at 120 hpf, the pancreatic expression persists, whereas the liver expression decreases below detection, and the gut expression decreases substantially, as well (Fig. 2E).
Effect of sbds gene knockdown.
To determine the role of the sbds gene perturbation on development, we performed gene knockdown using both translation blocking and splice blocking morpholinos (ATG, EI-1, and EI-2 respectively). We expected to see normally spliced transcript in wild type embryos and possibly an aberrant transcript in the EI-1 and EI-2 injected embryos due to the presence of introns. We obtained proof by RT-PCR showing the expected band in the control compared with numerous higher molecular weight species in the knockdown, likely due to the presence of introns (Fig. 3A). We sequenced the high molecular weight PCR product amplified from the EI-2 morpholino injected fish and proved that this is in fact the altered “splice” product. Conceptual translation of this product revealed an in-frame stop codon at position 88 (of 250 in the full-length product). This indicated that the aberrant transcript encodes a truncated protein. This suggests that we are effectively interfering with sbds gene expression. Inspection of live knockdown embryos up to 7 d after fertilization using a stereomicroscope showed no detectable developmental anomalies (Fig. 3B) and these embryos survive to juvenile stages. A likely explanation for this is the transience of morpholino gene knockdown, whereby wild-type gene product is expressed again at about 96–120 hpf.
Analysis of sbds morpholino knockdown embryos. (A) RT-PCR analysis of embryos injected with varying concentrations of EI-2 morpholino (0, 50, 100, 200 μM) at 24 and 48 hpf showing decreased product in injected embryos resulting in aberrant splice products (*). (B) Bright field images of live wild type and morpholino embryos at 7 d after fertilization showed no overt developmental anomalies. Scale bars: B, 250 μm.
Effect of sbds knockdown on pancreas development.
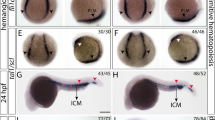
We performed ISH using the trypsin probe to stain the exocrine pancreas in 72 hpf larvae. Figure 4A,B shows the exocrine pancreas morphology in wild type and sbds morphant embryos. In the control group, we found that the exocrine pancreas completely surrounds the islets, consistent with previous reports (29). However, in the morphant group, we noticed that the exocrine tissue fails to completely surround the islets, leaving what appears to be a medial defect in the exocrine pancreas. We sectioned the embryos and showed that the exocrine tissue completely surrounds the islets in the control compared with a medial defect in the knockdown model (Fig. 4C,D). Interestingly, H&E sectioning of the wild type and knockdown embryos did not show any difference in acinar cell morphology and zymogen granule number or distribution (Fig. 4E,F). In addition, there was no discernible difference in the endocrine pancreas morphology based on staining for insulin between the wild type and morpholino embryos (Fig. 4G,H).
Effect of sbds knockdown on pancreatic development (A–H). Whole-mount in situ hybridizations showing zebrafish trypsin expression in wild type (A) and knockdown embryos (B). Cross section of 72 hpf wild type (C) and morpholino embryo (D) showing medial defect in the exocrine pancreas in the morpholino group. H&E staining showing normal exocrine cells and zymogen granule distribution in wild type (E) and morpholino embryo (F). Whole-mount in situ hybridizations showing zebrafish insulin expression in wild type (G) and morpholino embryo (H). Scale bars: A,B,G,H, 250 μm; C,D, 50 μm; E,F, 16 μm.
Role of sbds in neutrophil development.
To characterize the neutrophil development, we performed ISH using the myeloperoxidase (mpo) probe. Figure 5 shows the 48 hpf embryos in the lateral and ventral views. It has been shown that the neutrophil development starts in the posterior intermediate cell mass at 18 hpf and spreads anteriorly along the yolk sac (30). When we knock down the sbds gene, we noticed that the distribution is substantially different in the knockdown compared with wild type embryos. In the control, numerous mpo-positive cells are found across the surface of the yolk. In the sbds knockdown, there were virtually no mpo-positive cells detected on the yolk surface, although there was comparable staining for mpo in the location of the intermediate cell mass. Thus, it appears that the migration of granulocytes across the yolk surface may be impaired in the sbds knockdown model.
Effect of sbds knockdown on granulocyte development (A–F). (A,B) Dorsolateral view, with head to the left, (C–F) ventral view. Expression of mpo in wild type (A,C,E) and morpholino embryo (B,D,F) at 48 hpf. sbds knockdown embryos at 48 hpf lack mpo staining cells in the ventral view compared with wild type embryo. Scale bars: A–D, 250 μm; E,F, 100 μm.
DISCUSSION
This study was done to evaluate the pancreatic and myeloid development in a zebrafish model for the Shwachman-Diamond Syndrome. We identified and characterized the sbds gene in zebrafish. We observed that the sbds gene is expressed in zebrafish at different stages of development, from embryonic to larval. Whole-mount gene expression analysis revealed a dynamic pattern, in which sbds is expressed in and around the digestive organs. Histologic sectioning revealed sbds gene expression in the mesenchyme surrounding the digestive organs, with relatively less epithelial staining. When the sbds gene is knocked down with either start or splice morpholinos at the one-cell embryo stage, we observed an alteration in the spatial relationship between exocrine and endocrine pancreas such that the medial part of exocrine pancreas is absent. We also documented abnormal granulocytes distribution in the knockdown zebrafish model compared with the wild type embryos suggesting altered migration during development.
Individuals with Shwachman-Diamond syndrome have exocrine pancreatic insufficiency. This has been attributed to abnormal acinar development, and imaging and autopsy finding showed extensive fatty replacement of pancreatic acinar tissue with normal ductal and islet architecture. In contrast to the findings in humans, we noted normal acinar cell development, but rather a defect in the overall morphogenesis of the exocrine pancreas. This phenotype, taken together with the expression of sbds predominantly in the mesenchyme, suggests perhaps a loss of epithelial-mesenchymal interactions known to be necessary for proper exocrine development (31,32). Because there is no data on early pancreatic defects in humans with SDS, we can only speculate that an early defect in pancreatic morphogenesis could account for the pancreatic insufficiency seen in SDS patients. Fatty replacement of acinar tissue would then be a secondary consequence of an earlier defect. In addition, the morpholino knockdown is only transient; thus, our model describes only the early pancreatic acinar development. Because gene expression returns at 96 hpf, we cannot truly assess the long-term effect of loss of SBDS gene function in this model. Approaches that effect a more enduring loss of gene function at different stages of larval development may explain the long-term exocrine phenotype defect seen in patients with SD syndrome.
We also observed abnormal granulocytes distribution in the knockdown model, which is consistent with the abnormal chemotaxis defect seen in patients with Shwachman-diamond syndrome (33). To our knowledge, this is the first knockdown animal model (zebrafish) to show the pancreas and granulocytes developmental defect in Shwachman-Diamond syndrome. Previous attempts to delete or knockout Sbds gene resulted in lethality in S. cerevisiae and mouse, suggesting that this gene is essential for survival. Because knockdown in the zebrafish results in transient and partial loss of gene activity, this may explain why overall development proceeds without major flaws, enabling us to detect the developmental defect in the pancreas and granulocytes migration.
Previous studies in zebrafish have shown that the exocrine pancreas develops from the fusion of the ventral and dorsal buds at 72 hpf (29). Cell migration may thus play a role in exocrine pancreas morphogenesis. Likewise, migration is required for neutrophil function. Thus, we would speculate that the functional defects we see might be due to defective migration. An approach to test this hypothesis would be to use tissue-specific transgenic GFP (green fluorescent protein) fish to visualize the dynamic fusion of the dorsal and ventral pancreas defect in the knockdown model and to visualize neutrophil migration. Thus, the zebrafish model can be used to investigate which, and how, different organs are affected in Shwachman syndrome, making for a promising model system for future research and to develop novel therapies in this condition.
Abbreviations
- hpf:
-
hours post fertilization
- ISH:
-
in situ hybridization
- RT-PCR:
-
reverse transcriptase-polymerase chain reaction
- SDS:
-
Shwachman-Diamond Syndrome
- sbds :
-
Shwachman-Bodian-Diamond syndrome gene
References
Shwachman H, Diamond LK, Oski FA, Khaw KT 1964 The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr 65: 645–663
Bodian M, Sheldon W, Lightwood R 1964 Congenital hypoplasia of the exocrine pancreas. Acta Paediatr 53: 282–293
Ginzberg H, Shin J, Ellis L, Morrison J, Ip W, Dror Y, Freedman M, Heitlinger LA, Belt MA, Corey M, Rommens JM, Durie PR 1999 Shwachman syndrome: phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J Pediatr 135: 81–88
Makitie O, Ellis L, Durie PR, Morrison JA, Sochett EB, Rommens JM, Cole WG 2004 Skeletal phenotype in patients with Shwachman-Diamond syndrome and mutations in SBDS. Clin Genet 65: 101–112
Smith OP, Hann IM, Chessells JM, Reeves BR, Milla P 1996 Haematological abnormalities in Shwachman-Diamond syndrome. Br J Haematol 94: 279–284
Revert Lazaro F, Perez Monjardin E, Perez AP 2006 [Hypertransaminasemia as a manifestation of Shwachman-Diamond syndrome]. An Pediatr (Barc) 64: 481–484
Dror Y 2005 Shwachman-Diamond syndrome. Pediatr Blood Cancer 45: 892–901
Wilschanski M, van der Hoeven E, Phillips J, Shuckett B, Durie P 1994 Shwachman-Diamond syndrome presenting as hepatosplenomegaly. J Pediatr Gastroenterol Nutr 19: 111–113
Ritchie DS, Angus PW, Bhathal PS, Grigg AP 2002 Liver failure complicating non-alcoholic steatohepatitis following allogeneic bone marrow transplantation for Shwachman-Diamond syndrome. Bone Marrow Transplant 29: 931–933
Reif S, Arav-Boger R, Diamant S, Burstein Y, Fatal A 1999 Shwachman-Diamond syndrome associated with autoimmune phenomena. J Med 30: 259–265
Brueton MJ, Mavromichalis J, Goodchild MC, Anderson CM 1977 Hepatic dysfunction in association with pancreatic insufficiency and cyclical neutropenia. Shwachman-Diamond syndrome. Arch Dis Child 52: 76–78
Liebman WM, Rosental E, Hirshberger M, Thaler MM 1979 Shwachman-Diamond sydnrome and chronic liver disease. Clin Pediatr (Phila) 18: 695–696, 698
Aggett PJ, Cavanagh NP, Matthew DJ, Pincott JR, Sutcliffe J, Harries JT 1980 Shwachman's syndrome. A review of 21 cases. Arch Dis Child 55: 331–347
Savilahti E, Rapola J 1984 Frequent myocardial lesions in Shwachman's syndrome. Eight fatal cases among 16 Finnish patients. Acta Paediatr Scand 73: 642–651
Kent A, Murphy GH, Milla P 1990 Psychological characteristics of children with Shwachman syndrome. Arch Dis Child 65: 1349–1352
Ginzberg H, Shin J, Ellis L, Goobie S, Morrison J, Corey M, Durie PR, Rommens JM 2000 Segregation analysis in Shwachman-Diamond syndrome: evidence for recessive inheritance. Am J Hum Genet 66: 1413–1416
Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM 2003 Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33: 97–101
Goobie S, Popovic M, Morrison J, Ellis L, Ginzberg H, Boocock GR, Ehtesham N, Betard C, Brewer CG, Roslin NM, Hudson TJ, Morgan K, Fujiwara TM, Durie PR, Rommens JM 2001 Shwachman-Diamond syndrome with exocrine pancreatic dysfunction and bone marrow failure maps to the centromeric region of chromosome 7. Am J Hum Genet 68: 1048–1054
Popovic M, Goobie S, Morrison J, Ellis L, Ehtesham N, Richards N, Boocock G, Durie PR, Rommens JM 2002 Fine mapping of the locus for Shwachman-Diamond syndrome at 7q11, identification of shared disease haplotypes, and exclusion of TPST1 as a candidate gene. Eur J Hum Genet 10: 250–258
Shammas C, Menne TF, Hilcenko C, Michell SR, Goyenechea B, Boocock GR, Durie PR, Rommens JM, Warren AJ 2005 Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome. J Biol Chem 280: 19221–19229
Savchenko A, Krogan N, Cort JR, Evdokimova E, Lew JM, Yee AA, Sanchez-Pulido L, Andrade MA, Bochkarev A, Watson JD, Kennedy MA, Greenblatt J, Hughes T, Arrowsmith CH, Rommens JM, Edwards AM 2005 The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism. J Biol Chem 280: 19213–19220
Austin KM, Leary RJ, Shimamura A 2005 The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood 106: 1253–1258
Mack DR, Forstner GG, Wilschanski M, Freedman MH, Durie PR 1996 Shwachman syndrome: exocrine pancreatic dysfunction and variable phenotypic expression. Gastroenterology 111: 1593–1602
Bom EP, van der Sande FM, Tjon RT, Tham A, Hillen HF 1993 Shwachman syndrome: CT and MR diagnosis. J Comput Assist Tomogr 17: 474–476
MacMaster SA, Cummings TM 1993 Computed tomography and ultrasonography findings for an adult with Shwachman syndrome and pancreatic lipomatosis. Can Assoc Radiol J 44: 301–303
Zhang S, Shi M, Hui CC, Rommens JM 2006 Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol 26: 6656–6663
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995 Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310
Mayer AN, Fishman MC 2003 Nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development 130: 3917–3928
Field HA, Dong PD, Beis D, Stainier DY 2003 Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol 261: 197–208
Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT 2001 Myelopoiesis in the zebrafish, Danio rerio. Blood 98: 643–651
Kumar M, Jordan N, Melton D, Grapin-Botton A 2003 Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol 259: 109–122
Kumar M, Melton D 2003 Pancreas specification: a budding question. Curr Opin Genet Dev 13: 401–407
Stepanovic V, Wessels D, Goldman FD, Geiger J, Soll DR 2004 The chemotaxis defect of Shwachman-Diamond Syndrome leukocytes. Cell Motil Cytoskeleton 57: 158–174
Acknowledgements
We are grateful to Gigi Makky, Kate Marshall, Jian Zhang, and Amber Tomasini for generous assistance with experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by funds from the Children's Research Institute at Children's Hospital of Wisconsin, Milwaukee.
Rights and permissions
About this article
Cite this article
Venkatasubramani, N., Mayer, A. A Zebrafish Model for the Shwachman-Diamond Syndrome (SDS). Pediatr Res 63, 348–352 (2008). https://doi.org/10.1203/PDR.0b013e3181659736
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e3181659736
This article is cited by
-
Dual regulation of p53 by the ribosome maturation factor SBDS
Cell Death & Disease (2020)
-
Shwachman-Diamond Syndrome: Molecular Mechanisms and Current Perspectives
Molecular Diagnosis & Therapy (2019)