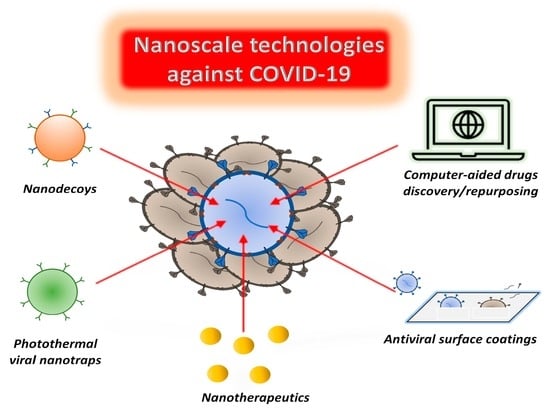
Nanoscale Technologies in the Fight against COVID-19: From Innovative Nanomaterials to Computer-Aided Discovery of Potential Antiviral Plant-Derived Drugs
Abstract
:1. Introduction
2. SARS-CoV-2 and COVID-19
3. Nanomaterials for COVID-19 Prevention and Treatment
3.1. Metal-Based Nanomaterials
3.1.1. Silver-Based Nanomaterials
3.1.2. Gold-Based Nanomaterials
3.1.3. Copper-Based Nanomaterials
3.1.4. Zinc-Based Nanomaterials
3.1.5. Nanomaterials Based on Other Transition Metals and Complex Metallic Alloys
3.2. Chitosan
3.3. Carbon- and Polymer-Based Nanomaterials
3.4. Cytomimetic Nanomaterials
3.5. Nanomaterials for Photothermal, Photodynamic, and Photocatalytic Treatment
3.6. DDSs for COVID-19 Therapy
4. Computer-Based Investigations for Discovery of Plant-Derived Drugs as Potential Anti-SARS-CoV-2 Agents: Focus on Terpenes and Terpenoids
4.1. Terpenes and Terpenoids as Antivirals
4.2. In Silico Studies on Terpenoids Interaction with ACE2 and SARS-CoV-2 Spike Protein
4.3. In Silico Studies on Terpenoids Interaction with GRP78 Protein
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin converting enzyme 2. |
| ADME | Absorption, Distribution, Metabolism, and Excretion. |
| aPDT | Antimicrobial photodynamic therapy. |
| ARDS | Acute respiratory distress syndrome. |
| ATP | Adenosine triphosphate. |
| AZT | Azithromycin. |
| BER | Berberin. |
| BSA | Bovine serum albumin. |
| BTB | Baricitinib. |
| CNP | 3-cyanopyridine. |
| Cys | Cysteine. |
| COVID | Coronavirus disease. |
| CT | Chitosan. |
| CTAB | Cetyltrimethylammonium bromide. |
| DHEA | Dehydroepiandrosterone. |
| DDSs | Drug delivery systems. |
| DNICs | Dinitrosyl iron complexes. |
| EPA | Environmental Protection Agency. |
| ER | Endoplasmic reticulum. |
| EUA | Emergency Use Authorization. |
| FDA | Food and Drug Administration. |
| FeO NPs | iron oxide nanoparticles. |
| GCPQ | N-palmitoyl-N-monomethyl-N,N-dimethyl-N,N,N-trimethyl-6-O-glycolchitosan. |
| GLU | Glutamate–urea-based ligand. |
| GO | Graphene oxide. |
| H2@C60 | Steroid–endohedral fullerene. |
| HEK | Human embryonic kidney. |
| His | Histidine. |
| HPVs | Human papillomaviruses. |
| HTCC | N-(2- hydroxypropyl)-3-trimethylammonium chitosan chloride. |
| IVM | Ivermectin. |
| LPH | Lipid polymer hybrid. |
| MERS | Middle-East Respiratory Syndrome. |
| MXenes | Transition metal carbides/nitrides. |
| Mpro | Main protease. |
| NCs | Nanocatchers. |
| NPs | Nanoparticles. |
| PDDA | Poly-diallyl-(dimethylammonium) chloride. |
| PLGA | Poly(lactic-co-glycolic acid). |
| PLpro | Papain-like protease. |
| PP | Polypropylene. |
| PSS | Poly (sodium 4-styrenesulfonate). |
| RBC | Red blood cell. |
| RBD | Receptor-binding domain. |
| RdRp | RNA-dependent RNA polymerase. |
| ROS | Reactive oxygen species. |
| RSV | Respiratory syncytial virus. |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2. |
| sgRNAs | Subgenomic RNAs. |
| SWCNTs | Single-walled carbon nanotubes. |
| Xenes | Metal carbides/nitrides. |
| 3CLpro | 3C-like proteinase. |
References
- World Health Organization Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 4 June 2022).
- Jayaweera, M.; Perera, H.; Gunawardana, B.; Manatunge, J. Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ. Res. 2020, 188, 109819. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.A.; Quandelacy, T.M.; Kada, S.; Prasad, P.V.; Steele, M.; Brooks, J.T.; Slayton, R.B.; Biggerstaff, M.; Butler, J.C. SARS-CoV-2 Transmission From People Without COVID-19 Symptoms. JAMA Netw. Open 2021, 4, e2035057. [Google Scholar] [CrossRef] [PubMed]
- Ravindra, K.; Malik, V.; Padhi, B.; Goel, S.; Gupta, M. Asymptomatic infection and transmission of COVID-19 among clusters: Systematic review and meta-analysis. Public Health 2022, 203, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Cimolai, N. In pursuit of the right tail for the COVID-19 incubation period. Public Health 2021, 194, 149–155. [Google Scholar] [CrossRef]
- Ma, Q.; Liu, J.; Liu, Q.; Kang, L.; Liu, R.; Jing, W.; Wu, Y.; Liu, M. Global Percentage of Asymptomatic SARS-CoV-2 Infections Among the Tested Population and Individuals With Confirmed COVID-19 Diagnosis. JAMA Netw. Open 2021, 4, e2137257. [Google Scholar] [CrossRef]
- Sah, P.; Fitzpatrick, M.C.; Zimmer, C.F.; Abdollahi, E.; Juden-Kelly, L.; Moghadas, S.M.; Singer, B.H.; Galvani, A.P. Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis. Proc. Natl. Acad. Sci. USA 2021, 118, e2109229118. [Google Scholar] [CrossRef]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe Covid-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Dessie, Z.G.; Zewotir, T. Mortality-related risk factors of COVID-19: A systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect. Dis. 2021, 21, 855. [Google Scholar] [CrossRef]
- Levy, J.H.; Iba, T.; Olson, L.B.; Corey, K.M.; Ghadimi, K.; Connors, J.M. COVID-19: Thrombosis, thromboinflammation, and anticoagulation considerations. Int. J. Lab. Hematol. 2021, 43, 29–35. [Google Scholar] [CrossRef]
- Shafqat, A.; Shafqat, S.; Salameh, S.A.; Kashir, J.; Alkattan, K.; Yaqinuddin, A. Mechanistic Insights Into the Immune Pathophysiology of COVID-19; An In-Depth Review. Front. Immunol. 2022, 13, 835104. [Google Scholar] [CrossRef]
- Singh, L.; Kruger, H.G.; Maguire, G.E.; Govender, T.; Parboosing, R. The role of nanotechnology in the treatment of viral infections. Ther. Adv. Infect. Dis. 2017, 4, 105–131. [Google Scholar] [CrossRef]
- Rashidzadeh, H.; Danafar, H.; Rahimi, H.; Mozafari, F.; Salehiabar, M.; Rahmati, M.A.; Rahamooz-Haghighi, S.; Mousazadeh, N.; Mohammadi, A.; Ertas, Y.N.; et al. Nanotechnology against the novel coronavirus (severe acute respiratory syndrome coronavirus 2): Diagnosis, treatment, therapy and future perspectives. Nanomedicine 2021, 16, 497–516. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, G.; La, M.; Liu, L. Colorimetric and Electrochemical Methods for the Detection of SARS-CoV-2 Main Protease by Peptide-Triggered Assembly of Gold Nanoparticles. Molecules 2022, 27, 615. [Google Scholar] [CrossRef]
- Lim, J.W.; Ahn, Y.R.; Park, G.; Kim, H.O.; Haam, S. Application of Nanomaterials as an Advanced Strategy for the Diagnosis, Prevention, and Treatment of Viral Diseases. Pharmaceutics 2021, 13, 1570. [Google Scholar] [CrossRef]
- Gunnels, T.F.; Stranford, D.M.; Mitrut, R.E.; Kamat, N.P.; Leonard, J.N. Elucidating Design Principles for Engineering Cell-Derived Vesicles to Inhibit SARS-CoV-2 Infection. Small 2022, 18, 2200125. [Google Scholar] [CrossRef]
- Liang, L.; Ahamed, A.; Ge, L.; Fu, X.; Lisak, G. Advances in Antiviral Material Development. ChemPlusChem 2020, 85, 2105–2128. [Google Scholar] [CrossRef]
- Tavakoli, A.; Hashemzadeh, M.S. Inhibition of herpes simplex virus type 1 by copper oxide nanoparticles. J. Virol. Methods 2020, 275, 113688. [Google Scholar] [CrossRef]
- Ghaffari, H.; Tavakoli, A.; Moradi, A.; Tabarraei, A.; Bokharaei-Salim, F.; Zahmatkeshan, M.; Farahmand, M.; Javanmard, D.; Kiani, S.J.; Esghaei, M.; et al. Inhibition of H1N1 influenza virus infection by zinc oxide nanoparticles: Another emerging application of nanomedicine. J. Biomed. Sci. 2019, 26, 70. [Google Scholar] [CrossRef]
- Yang, X.X.; Li, C.M.; Huang, C.Z. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale 2016, 8, 3040–3048. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, W.; Wang, W.; Zhang, X.; Zhao, X. The inhibitory effects and mechanisms of 3,6-O-sulfated chitosan against human papillomavirus infection. Carbohydr. Polym. 2018, 198, 329–338. [Google Scholar] [CrossRef]
- Motelica, L.; Ficai, D.; Ficai, A.; Truşcă, R.D.; Ilie, C.I.; Oprea, O.C.; Andronescu, E. Innovative Antimicrobial Chitosan/ZnO/Ag NPs/Citronella Essential Oil Nanocomposite—Potential Coating for Grapes. Foods 2020, 9, 1801. [Google Scholar] [CrossRef] [PubMed]
- Villa-Hermosilla, M.C.; Negro, S.; Barcia, E.; Hurtado, C.; Montejo, C.; Alonso, M.; Fernandez-Carballido, A. Celecoxib Microparticles for Inhalation in COVID-19-Related Acute Respiratory Distress Syndrome. Pharmaceutics 2022, 14, 1392. [Google Scholar] [CrossRef] [PubMed]
- Bentley, K.; Stanton, R.J. Hydroxypropyl Methylcellulose-Based Nasal Sprays Effectively Inhibit In Vitro SARS-CoV-2 Infection and Spread. Viruses 2021, 13, 2345. [Google Scholar] [CrossRef] [PubMed]
- Mullins, J.G. Drug repurposing Silico Screen. Platforms. Biochem. Soc. Trans. 2022, 50, 747–758. [Google Scholar] [CrossRef]
- Ma, L.; Li, H.; Lan, J.; Hao, X.; Liu, H.; Wang, X.; Huang, Y. Comprehensive analyses of bioinformatics applications in the fight against COVID-19 pandemic. Comput. Biol. Chem. 2021, 95, 107599. [Google Scholar] [CrossRef]
- Khan, M.F.; Ansari, W.A.; Ahamad, T.; Khan, M.A.; Khan, Z.A.; Sarfraz, A.; Khan, M.A. Bioactive components of different nasal spray solutions may defeat SARS-Cov2: Repurposing and in silico studies. J. Mol. Model. 2022, 28, 212. [Google Scholar] [CrossRef]
- Al-Sanea, M.M.; Abelyan, N.; Abdelgawad, M.A.; Musa, A.; Ghoneim, M.M.; Al-Warhi, T.; Aljaeed, N.; Alotaibi, O.J.; Alnusaire, T.S.; Abdelwahab, S.F.; et al. Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling. Antibiotics 2021, 10, 824. [Google Scholar] [CrossRef]
- Eissa, I.H.; Alesawy, M.S.; Saleh, A.M.; Elkaeed, E.B.; Alsfouk, B.A.; El-Attar, A.A.M.M.; Metwaly, A.M. Ligand and Structure-Based In Silico Determination of the Most Promising SARS-CoV-2 nsp16-nsp10 2′-o-Methyltransferase Complex Inhibitors among 3009 FDA Approved Drugs. Molecules 2022, 27, 2287. [Google Scholar] [CrossRef]
- Refat, M.S.; Bakare, S.B.; Altalhi, T.A.; Alam, K.; Al-Hazmi, G.H. Synthesis and spectroscopic interpretations of Co(II), Ni(II) and Cu(II) decxycholate complexes with molecular docking of COVID-19 protease. Pol. J. Chem. Technol. 2021, 23, 54–59. [Google Scholar] [CrossRef]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef]
- Huang, J.; Tao, G.; Liu, J.; Cai, J.; Huang, Z.; Chen, X.-J. Current Prevention of COVID-19: Natural Products and Herbal Medicine. Front. Pharmacol. 2020, 11, 588508. [Google Scholar] [CrossRef]
- Islam, F.; Bibi, S.; Meem, A.F.K.; Islam, M.M.; Rahaman, M.S.; Bepary, S.; Rahman, M.M.; Rahman, M.M.; Elzaki, A.; Kajoak, S.; et al. Natural Bioactive Molecules: An Alternative Approach to the Treatment and Control of COVID-19. Int. J. Mol. Sci. 2021, 22, 12638. [Google Scholar] [CrossRef]
- Ma, L.L.; Liu, H.M.; Liu, X.M.; Yuan, X.Y.; Xu, C.; Wang, F.; Lin, J.Z.; Xu, R.C.; Zhang, D.K. Screening S protein—ACE2 blockers from natural products: Strategies and advances in the discovery of potential inhibitors of COVID-19. Eur. J. Med. Chem. 2021, 226, 113857. [Google Scholar] [CrossRef]
- Soleymani, S.; Naghizadeh, A.; Karimi, M.; Zarei, A.; Mardi, R.; Kordafshari, G.; Esmaealzadeh, N.; Zargaran, A. COVID-19: General Strategies for Herbal Therapies. J. Evid.-Based Integr. Med. 2022, 27, 2515690X2110536. [Google Scholar] [CrossRef]
- Motelica, L.; Ficai, D.; Oprea, O.C.; Ficai, A.; Ene, V.L.; Vasile, B.S.; Andronescu, E.; Holban, A.M. Antibacterial Biodegradable Films Based on Alginate with Silver Nanoparticles and Lemongrass Essential Oil–Innovative Packaging for Cheese. Nanomaterials 2021, 11, 2377. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Mashrur, F.R.; Chhoan, A.P.; Shahriar, S.M.; Haidere, M.F.; Runa, N.J.; Kim, S.; Kweon, D.H.; Hosseinzadeh, H.; Cho, J.Y. Silver Nanoparticles as Potential Antiviral Agents. Pharmaceutics 2021, 13, 2034. [Google Scholar] [CrossRef]
- Chircov, C.; Matei, M.F.; Neacșu, I.A.; Vasile, B.S.; Oprea, O.C.; Croitoru, A.M.; Trușcă, R.D.; Andronescu, E.; Sorescu, I.; Bărbuceanu, F. Iron Oxide–Silica Core–Shell Nanoparticles Functionalized with Essential Oils for Antimicrobial Therapies. Antibiotics 2021, 10, 1138. [Google Scholar] [CrossRef]
- Mallakpour, S.; Azadi, E.; Hussain, C.M. The latest strategies in the fight against the COVID-19 pandemic: The role of metal and metal oxide nanoparticles. New J. Chem. 2021, 45, 6167–6179. [Google Scholar] [CrossRef]
- Motelica, L.; Popescu, A.; Răzvan, A.G.; Oprea, O.; Truşcă, R.D.; Vasile, B.S.; Dumitru, F.; Holban, A.M. Facile Use of ZnO Nanopowders to Protect Old Manual Paper Documents. Materials 2020, 13, 5452. [Google Scholar] [CrossRef]
- Kiel, A.; Kaltschmidt, B.P.; Asghari, E.; Hütten, A.; Kaltschmidt, B.; Kaltschmidt, C. Bacterial Biofilm Formation on Nano-Copper Added PLA Suited for 3D Printed Face Masks. Microorganisms 2022, 10, 439. [Google Scholar] [CrossRef]
- Purniawan, A.; Lusida, M.I.; Pujiyanto, R.W.; Nastri, A.M.; Permanasari, A.A.; Harsono, A.A.H.; Oktavia, N.H.; Wicaksono, S.T.; Dewantari, J.R.; Prasetya, R.R.; et al. Synthesis and assessment of copper-based nanoparticles as a surface coating agent for antiviral properties against SARS-CoV-2. Sci. Rep. 2022, 12, 4835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, W.; Sun, Y.; Chen, W.; Zhang, Y. Application of antiviral materials in textiles: A review. Nanotechnol. Rev. 2021, 10, 1092–1115. [Google Scholar] [CrossRef]
- Kampf, G. How long can nosocomial pathogens survive on textiles? A systematic review. GMS Hyg Infect Control. 2020, 15, Doc10. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C.; et al. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef] [PubMed]
- Bisht, D.; Rashid, M.; Arya, R.K.K.; Kumar, D.; Chaudhary, S.K.; Rana, V.S.; Sethiya, N.K. Revisiting liquorice (Glycyrrhiza glabra L.) as anti-inflammatory, antivirals and immunomodulators: Potential pharmacological applications with mechanistic insight. Phytomedicine Plus 2022, 2, 100206. [Google Scholar] [CrossRef]
- Pollard, B.S.; Blancol, J.C.; Pollard, J.R. Classical Drug Digitoxin Inhibits Influenza Cytokine Storm, With Implications for Covid-19 Therapy. In Vivo 2020, 34, 3723–3730. [Google Scholar] [CrossRef]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus Infections—More Than Just the Common Cold. JAMA 2020, 323, 707. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2021, 23, 3–20. [Google Scholar] [CrossRef]
- Ebner, B.; Volz, Y.; Mumm, J.N.; Stief, C.G.; Magistro, J. The COVID-19 pandemic—What have urologists learned? Nat. Rev. Urol. 2022, 19, 344–356. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Suryamohan, K.; Diwanji, D.; Stawiski, E.W.; Gupta, R.; Miersch, S.; Liu, J.; Chen, C.; Jiang, Y.P.; Fellouse, F.A.; Sathirapongsasuti, J.F.; et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 2021, 4, 475. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Raghav, P.K.; Kalyanaraman, K.; Kumar, D. Human cell receptors: Potential drug targets to combat COVID-19. Amino Acids 2021. [Google Scholar] [CrossRef]
- Wang, M.; Wey, S.; Zhang, Y.; Ye, R.; Lee, A.S. Role of the Unfolded Protein Response Regulator GRP78/BiP in Development, Cancer, and Neurological Disorders. Antioxid. Redox Signal. 2009, 11, 2307–2316. [Google Scholar] [CrossRef]
- Palmeira, A.; Sousa, E.; Köseler, A.; Sabirli, R.; Gören, T.; Türkçüer, İ.; Kurt, O.; Pinto, M.M.; Vasconcelos, M.H. Preliminary Virtual Screening Studies to Identify GRP78 Inhibitors Which May Interfere with SARS-CoV-2 Infection. Pharmaceuticals 2020, 13, 132. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elfiky, A.A. GRP78: A cell’s response to stress. Life Sci. 2019, 226, 156–163. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, S.; Disoma, C.; Zheng, R.; Zhou, M.; Razzaq, A.; Liu, P.; Zhou, Y.; Dong, Z.; Du, A.; et al. SARS-CoV-2: Mechanism of infection and emerging technologies for future prospects. Rev. Med. Virol. 2020, 31, e2168. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.A.; Lee, K.M.; Chiang, H.J.; Huang, S.Y.; Wu, C.J.; Shih, S.R. Molecular Virology of SARS-CoV-2 and Related Coronaviruses. Microbiol. Mol. Biol. Rev. 2022, 86, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.; Roustit, M.; Lepelley, M.; Revol, B.; Cracowski, J.L.; Khouri, C. Reported Adverse Drug Reactions Associated With the Use of Hydroxychloroquine and Chloroquine During the COVID-19 Pandemic. Ann. Intern. Med. 2021, 174, 878–880. [Google Scholar] [CrossRef]
- Roustit, M.; Guilhaumou, R.; Molimard, M.; Drici, M.D.; Laporte, S.; Montastruc, J.L. Chloroquine and hydroxychloroquine in the management of COVID-19: Much kerfuffle but little evidence. Therapies 2020, 75, 363–370. [Google Scholar] [CrossRef]
- Garegnani, L.I.; Madrid, E.; Meza, N. Misleading clinical evidence and systematic reviews on ivermectin for COVID-19. BMJ Evid.-Based Med. 2021, 27, 156–158. [Google Scholar] [CrossRef]
- Bryant, A.; Lawrie, T.A.; Dowswell, T.; Fordham, E.J.; Mitchell, S.; Hill, S.R.; Tham, T.C. Ivermectin for Prevention and Treatment of COVID-19 Infection. Am. J. Ther. 2021, 28, e434–e460. [Google Scholar] [CrossRef]
- van de Veerdonk, F.L.; Giamarellos-Bourboulis, E.; Pickkers, P.; Derde, L.; Leavis, H.; van Crevel, R.; Engel, J.J.; Wiersinga, W.J.; Vlaar, A.P.J.; Shankar-Hari, M.; et al. A guide to immunotherapy for COVID-19. Nat. Med. 2022, 28, 39–50. [Google Scholar] [CrossRef]
- Pelaia, C.; Calabrese, C.; Garofalo, E.; Bruni, A.; Vatrella, A.; Pelaia, G. Therapeutic Role of Tocilizumab in SARS-CoV-2-Induced Cytokine Storm: Rationale and Current Evidence. Int. J. Mol. Sci. 2021, 22, 3059. [Google Scholar] [CrossRef]
- Abidi, E.; Nekidy, W.S.E.; Alefishat, E.; Rahman, N.; Petroianu, G.A.; El-Lababidi, R.; Mallat, J. Tocilizumab and COVID-19: Timing of Administration and Efficacy. Front. Pharmacol. 2022, 13, 825749. [Google Scholar] [CrossRef]
- Interleukin-6 Inhibitors|COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/immunomodulators/interleukin-6-inhibitors/ (accessed on 4 June 2022).
- RoActemra|European Medicines Agency. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/roactemra (accessed on 8 January 2022).
- Popp, M.; Stegemann, M.; Riemer, M.; Metzendorf, M.I.; Romero, C.S.; Mikolajewska, A.; Kranke, P.; Meybohm, P.; Skoetz, N.; Weibel, S. Antibiotics for the treatment of COVID-19. Cochrane Database Syst. Rev. 2021, 2021, 1–124. [Google Scholar] [CrossRef]
- Conzelmann, C.; Müller, J.A.; Perkhofer, L.; Sparrer, K.M.; Zelikin, A.N.; Münch, J.; Kleger, A. Inhaled and systemic heparin as a repurposed direct antiviral drug for prevention and treatment of COVID-19. Clin. Med. 2020, 20, e218–e221. [Google Scholar] [CrossRef]
- Cuker, A.; Tseng, E.K.; Nieuwlaat, R.; Angchaisuksiri, P.; Blair, C.; Dane, K.; Davila, J.; DeSancho, M.T.; Diuguid, D.; Griffin, D.O.; et al. American Society of Hematology 2021 guidelines on the use of anticoagulation for thromboprophylaxis in patients with COVID-19. Blood Adv. 2021, 5, 872–888. [Google Scholar] [CrossRef]
- Eastman, R.T.; Roth, J.S.; Brimacombe, K.R.; Simeonov, A.; Shen, M.; Patnaik, S.; Hall, M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020, 6, 672–683. [Google Scholar] [CrossRef]
- Veklury|European Medicines Agency. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/veklury (accessed on 4 June 2022).
- Remdesivir|COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/remdesivir/ (accessed on 4 June 2022).
- Lamb, Y.N. Nirmatrelvir Plus Ritonavir: First Approval. Drugs 2022, 82, 585–591. [Google Scholar] [CrossRef]
- Syed, Y.Y. Molnupiravir: First Approval. Drugs 2022, 82, 455–460. [Google Scholar] [CrossRef]
- Anti-SARS-CoV-2 Monoclonal Antibodies|COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/anti-sars-cov-2-antibody-products/anti-sars-cov-2-monoclonal-antibodies/ (accessed on 4 June 2022).
- Ronapreve|COVID-19 Treatment Guidelines. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/ronapreve (accessed on 4 June 2022).
- Xevudy|COVID-19 Treatment Guidelines. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/xevudy (accessed on 4 June 2022).
- Patoo, T.S.; Khanday, F.; Qurashi, A. Prospectus of advanced nanomaterials for antiviral properties. Mater. Adv. 2022, 3, 2960–2970. [Google Scholar] [CrossRef]
- Cramer, C. Über Oligodynamische Erscheinungen in Lebenden Zellen von C. von Nägeli; Allgemeine Schweizerische Gesellschaft, &c. Neue Denkschriften, &c Tom. XXXIII, livr. 1: 1893. Available online: https://books.google.com.hk/books/about/%C3%9Cber_oligodynamische_Erscheinungen_in_l.html?id=o2xImgEACAAJ&redir_esc=y (accessed on 15 June 2022).
- Cirri, D.; Marzo, T.; Tolbatov, I.; Marrone, A.; Saladini, F.; Vicenti, I.; Dragoni, F.; Boccuto, A.; Messori, L. In Vitro Anti-SARS-CoV-2 Activity of Selected Metal Compounds and Potential Molecular Basis for Their Actions Based on Computational Study. Biomolecules 2021, 11, 1858. [Google Scholar] [CrossRef]
- Sportelli, M.C.; Izzi, M.; Kukushkina, E.A.; Hossain, S.I.; Picca, R.A.; Ditaranto, N.; Cioffi, N. Can Nanotechnology and Materials Science Help the Fight against SARS-CoV-2? Nanomaterials 2020, 10, 802. [Google Scholar] [CrossRef]
- Merkl, P.; Long, S.; McInerney, G.M.; Sotiriou, G.A. Antiviral Activity of Silver, Copper Oxide and Zinc Oxide Nanoparticle Coatings against SARS-CoV-2. Nanomaterials 2021, 11, 1312. [Google Scholar] [CrossRef]
- Gherasim, O.; Puiu, R.A.; Bîrcă, A.C.; Burdușel, A.C.; Grumezescu, A.M. An Updated Review on Silver Nanoparticles in Biomedicine. Nanomaterials 2020, 10, 2318. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Deshmukh, S.D.; Ingle, A.P.; Gupta, I.R.; Galdiero, M.; Galdiero, S. Metal nanoparticles: The protective nanoshield against virus infection. Crit. Rev. Microbiol. 2014, 42, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Elechiguerra, J.L.; Burt, J.L.; Morones, J.R.; Camacho-Bragado, A.; Gao, X.; Lara, H.H.; Yacaman, M.J. Interaction of silver nanoparticles with HIV-1. J. Nanobiotechnology 2005, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.S.M.; Rodrigues, A.M.; do Nascimento Souza, D.; de Novais, E.R.P.; Rodrigues, A.M.; de Oliveira, G.C.A.; de Lima Ferreira Novais, A. DFT calculations to investigate silver ions as a virucide from SARS-CoV-2. J. Mol. Model. 2021, 27, 323. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, S.S.; Miyakawa, K.; Morita, T.; Yamaoka, Y.; Ryo, A. Potent antiviral effect of silver nanoparticles on SARS-CoV-2. Biochem. Biophys. Res. Commun. 2020, 533, 195–200. [Google Scholar] [CrossRef]
- Babaei, A.; Mousavi, S.M.; Ghasemi, M.; Pirbonyeh, N.; Soleimani, M.; Moattari, A. Gold nanoparticles show potential in vitro antiviral and anticancer activity. Life Sci. 2021, 284, 119652. [Google Scholar] [CrossRef]
- Rothan, H.A.; Stone, S.; Natekar, J.; Kumari, P.; Arora, K.; Kumar, M. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-COV-2) replication and attenuates inflammation in human cells. Virology 2020, 547, 7–11. [Google Scholar] [CrossRef]
- Gil-Moles, M.; Basu, U.; Büssing, R.; Hoffmeister, H.; Türck, S.; Varchmin, A.; Ott, I. Gold Metallodrugs to Target Coronavirus Proteins: Inhibitory Effects on the Spike-ACE2 Interaction and on PLpro Protease Activity by Auranofin and Gold Organometallics. Chem. A Eur. J. 2020, 26, 15140–15144. [Google Scholar] [CrossRef]
- Mehranfar, A.; Izadyar, M. Theoretical Design of Functionalized Gold Nanoparticles as Antiviral Agents against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Phys. Chem. Lett. 2020, 11, 10284–10289. [Google Scholar] [CrossRef]
- Mertens, B.S.; Moore, M.D.; Jaykus, L.A.; Velev, O.D. Efficacy and Mechanisms of Copper Ion-Catalyzed Inactivation of Human Norovirus. ACS Infect. Dis. 2022, 8, 855–864. [Google Scholar] [CrossRef]
- Vincent, M.; Duval, R.; Hartemann, P.; Engels-Deutsch, M. Contact killing and antimicrobial properties of copper. J. Appl. Microbiol. 2018, 124, 1032–1046. [Google Scholar] [CrossRef]
- Hang, X.; Peng, H.; Song, H.; Qi, Z.; Miao, X.; Xu, W. Antiviral activity of cuprous oxide nanoparticles against Hepatitis C Virus in vitro. J. Virol. Methods 2015, 222, 150–157. [Google Scholar] [CrossRef]
- Aallaei, M.; Molaakbari, E.; Mostafavi, P.; Salarizadeh, N.; Maleksah, R.E.; Afzali, D. Investigation of Cu metal nanoparticles with different morphologies to inhibit SARS-CoV-2 main protease and spike glycoprotein using Molecular Docking and Dynamics Simulation. J. Mol. Struct. 2022, 1253, 132301. [Google Scholar] [CrossRef]
- Takeda, Y.; Jamsransuren, D.; Matsuda, S.; Crea, R.; Ogawa, H. The SARS-CoV-2-Inactivating Activity of Hydroxytyrosol-Rich Aqueous Olive Pulp Extract (HIDROX®) and Its Use as a Virucidal Cream for Topical Application. Viruses 2021, 13, 232. [Google Scholar] [CrossRef]
- Masarwa, M.; Cohen, H.; Meyerstein, D.; Hickman, D.L.; Bakac, A.; Espenson, J.H. Reactions of low-valent transition-metal complexes with hydrogen peroxide. Are they “Fenton-like” or not? 1. The case of + and . J. Am. Chem. Soc. 1988, 110, 4293–4297. [Google Scholar] [CrossRef]
- Jung, S.; Yang, J.Y.; Byeon, E.Y.; Kim, D.G.; Lee, D.G.; Ryoo, S.; Lee, S.; Shin, C.W.; Jang, H.W.; Kim, H.J.; et al. Copper-Coated Polypropylene Filter Face Mask with SARS-CoV-2 Antiviral Ability. Polymers 2021, 13, 1367. [Google Scholar] [CrossRef]
- Massaro, M.; Noto, R.; Riela, S. Past, Present and Future Perspectives on Halloysite Clay Minerals. Molecules 2020, 25, 4863. [Google Scholar] [CrossRef]
- Awad, M.E.; López-Galindo, A.; Setti, M.; El-Rahmany, M.M.; Iborra, C.V. Kaolinite in pharmaceutics and biomedicine. Int. J. Pharm. 2017, 533, 34–48. [Google Scholar] [CrossRef]
- Rius-Rocabert, S.; Arranz-Herrero, J.; Fernández-Valdés, A.; Marciello, M.; Moreno, S.; Llinares-Pinel, F.; Presa, J.; Hernandez-Alcoceba, R.; López-Píriz, R.; Torrecillas, R.; et al. Broad virus inactivation using inorganic micro/nano-particulate materials. Mater. Today Bio 2022, 13, 100191. [Google Scholar] [CrossRef]
- Almalki, S.A.; Bawazeer, T.M.; Asghar, B.; Alharbi, A.; Aljohani, M.M.; Khalifa, M.E.; El-Metwaly, N. Synthesis and characterization of new thiazole-based Co(II) and Cu(II) complexes; therapeutic function of thiazole towards COVID-19 in comparing to current antivirals in treatment protocol. J. Mol. Struct. 2021, 1244, 130961. [Google Scholar] [CrossRef]
- Gammoh, N.; Rink, L. Zinc in Infection and Inflammation. Nutrients 2017, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Kochańczyk, T.; Drozd, A.; Krężel, A. Relationship between the architecture of zinc coordination and zinc binding affinity in proteins – insights into zinc regulation. Metallomics 2015, 7, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Faten, F.; Ibrahim, S.R. Comparing Surface Chemical Modifications of Zinc Oxide Nanoparticles for Modulating their Antiviral Activity against Herpes Simplex Virus Type-1. Int. J. Nanoparticles Nanotechnol. 2018, 4, 1–14. [Google Scholar] [CrossRef]
- Read, S.A.; Obeid, S.; Ahlenstiel, C.; Ahlenstiel, G. The Role of Zinc in Antiviral Immunity. Adv. Nutr. 2019, 10, 696–710. [Google Scholar] [CrossRef] [PubMed]
- do Nascimento Marreiro, D.; Cruz, K.J.C.; de Oliveira, A.R.S.; Morais, J.B.S.; de Jesus e Silva de Almendra Freitas, B.; de Sousa Melo, S.R.; dos Santos, L.R.; Cardoso, B.E.P.; da Silva Dias, T.M. Antiviral and immunological activity of zinc and possible role in COVID-19. Br. J. Nutr. 2021, 127, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Gopal, V.; Nilsson-Payant, B.E.; French, H.; Siegers, J.Y.; Yung, W.-S.; Hardwick, M.; te Velthuis, A.J.W. Zinc-Embedded Polyamide Fabrics Inactivate SARS-CoV-2 and Influenza A Virus. ACS Appl. Mater. Interfaces 2021, 13, 30317–30325. [Google Scholar] [CrossRef]
- Hosseini, M.; Behzadinasab, S.; Chin, A.W.; Poon, L.L.; Ducker, W.A. Reduction of Infectivity of SARS-CoV-2 by Zinc Oxide Coatings. ACS Biomater. Sci. Eng. 2021, 7, 5022–5027. [Google Scholar] [CrossRef]
- Pormohammad, A.; Monych, N.; Turner, R. Zinc and SARS-CoV-2: A molecular modeling study of Zn interactions with RNA-dependent RNA-polymerase and 3C-like proteinase enzymes. Int. J. Mol. Med. 2020, 47, 326–334. [Google Scholar] [CrossRef]
- Panchariya, L.; Khan, W.A.; Kuila, S.; Sonkar, K.; Sahoo, S.; Ghoshal, A.; Kumar, A.; Verma, D.K.; Hasan, A.; Khan, M.A.; et al. Zinc2+ ion inhibits SARS-CoV-2 main protease and viral replication. Vitr. Chem. Commun. 2021, 57, 10083–10086. [Google Scholar] [CrossRef]
- Grifagni, D.; Calderone, V.; Junntini, S.; Cantini, F.; Fragai, M.; Banci, L. SARS-CoV-2 Mpro inhibition by a zinc ion: Structural features and hints for drug design. Chem. Commun. 2021, 57, 7910–7913. [Google Scholar] [CrossRef]
- Tao, X.; Zhang, L.; Du, L.; Lu, K.; Zhao, Z.; Xie, Y.; Li, X.; Huang, S.; Wang, P.H.; Pan, J.A.; et al. Inhibition of SARS-CoV-2 replication by zinc gluconate in combination with hinokitiol. J. Inorg. Biochem. 2022, 231, 111777. [Google Scholar] [CrossRef]
- Hamdi, M.; Abdel-Bar, H.M.; Elmowafy, E.; El-khouly, A.; Mansour, M.; Awad, G.A. Investigating the Internalization and COVID-19 Antiviral Computational Analysis of Optimized Nanoscale Zinc Oxide. ACS Omega 2021, 6, 6848–6860. [Google Scholar] [CrossRef]
- Adhikari, A.; Pal, U.; Bayan, S.; Mondal, S.; Ghosh, R.; Darbar, S.; Saha-Dasgupta, T.; Ray, S.K.; Pal, S.K. Nanoceutical Fabric Prevents COVID-19 Spread through Expelled Respiratory Droplets: A Combined Computational, Spectroscopic, and Antimicrobial Study. ACS Appl. Bio Mater. 2021, 4, 5471–5484. [Google Scholar] [CrossRef]
- Sportelli, M.C.; Izzi, M.; Loconsole, D.; Sallustio, A.; Picca, R.A.; Felici, R.; Chironna, M.; Cioffi, N. On the Efficacy of ZnO Nanostructures against SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 3040. [Google Scholar] [CrossRef]
- El-Megharbel, S.M.; Alsawat, M.; Al-Salmi, F.A.; Hamza, R.Z. Utilizing of (Zinc Oxide Nano-Spray) for Disinfection against “SARS-CoV-2” and Testing Its Biological Effectiveness on Some Biochemical Parameters during (COVID-19 Pandemic)—”ZnO Nanoparticles Have Antiviral Activity against (SARS-CoV-2)”. Coatings 2021, 11, 388. [Google Scholar] [CrossRef]
- Öztürkkan, F.E.; Özdemir, M.; Akbaba, G.B.; Sertçelik, M.; Yalçın, B.; Necefoğlu, H.; Hökelek, T. Synthesis, crystal structure, potential drug properties for Coronavirus of Co(II) and Zn(II) 2-chlorobenzoate with 3-cyanopyridine complexes. J. Mol. Struct. 2022, 1250, 131825. [Google Scholar] [CrossRef]
- Kumar, R.; Nayak, M.; Sahoo, G.C.; Pandey, K.; Sarkar, M.C.; Ansari, Y.; Das, V.; Topno, R.; Bhawna; Madhukar, M.; et al. Iron oxide nanoparticles based antiviral activity of H1N1 influenza A virus. J. Infect. Chemother. 2019, 25, 325–329. [Google Scholar] [CrossRef]
- Abo-zeid, Y.; Ismail, N.S.; McLean, G.R.; Hamdy, N.M. A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur. J. Pharm. Sci. 2020, 153, 105465. [Google Scholar] [CrossRef]
- Pectol, D.C.; DeLaney, C.R.; Zhu, J.; Mellott, D.M.; Katzfuss, A.; Taylor, Z.W.; Meek, T.D.; Darensbourg, M.Y. Dinitrosyl iron complexes (DNICs) as inhibitors of the SARS-CoV-2 main protease. Chem. Commun. 2021, 57, 8352–8355. [Google Scholar] [CrossRef]
- Mohamed, G.G.; Omar, M.M.; Ahmed, Y.M. Metal complexes of Tridentate Schiff base: Synthesis, Characterization, Biological Activity and Molecular Docking Studies with COVID-19 Protein Receptor. Z. Anorg. Allg. Chem. 2021, 647, 2201–2218. [Google Scholar] [CrossRef]
- Toledo, G.G.D.; Toledo, V.H.; Lanfredi, A.J.; Escote, M.; Champi, A.; Silva, M.C.C.D.; Nantes-Cardoso, I.L. Promising Nanostructured Materials against Enveloped Virus. An. Acad. Bras. Ciências 2020, 92, e20200718. [Google Scholar] [CrossRef] [PubMed]
- Hamza, R.Z.; Gobouri, A.A.; Al-Yasi, H.M.; Al-Talhi, T.A.; El-Megharbel, S.M. A New Sterilization Strategy Using TiO2 Nanotubes for Production of Free Radicals that Eliminate Viruses and Application of a Treatment Strategy to Combat Infections Caused by Emerging SARS-CoV-2 during the COVID-19 Pandemic. Coatings 2021, 11, 680. [Google Scholar] [CrossRef]
- Unal, M.A.; Bayrakdar, F.; Fusco, L.; Besbinar, O.; Shuck, C.E.; Yalcin, S.; Erken, M.T.; Ozkul, A.; Gurcan, C.; Panatli, O.; et al. 2D MXenes with antiviral and immunomodulatory properties: A pilot study against SARS-CoV-2. Nano Today 2021, 38, 101136. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Phadke, K.S.; Li, Z.; Ouyang, G.; hoon Kim, T.; Zhou, L.; Slaughter, J.; Bellaire, B.; Ren, S.; Cui, J. Sprayable copper and copper–zinc nanowires inks for antiviral surface coating. RSC Adv. 2022, 12, 6093–6098. [Google Scholar] [CrossRef]
- Zhou, Y.; Fletcher, N.F.; Zhang, N.; Hassan, J.; Gilchrist, M.D. Enhancement of Antiviral Effect of Plastic Film against SARS-CoV-2: Combining Nanomaterials and Nanopatterns with Scalability for Mass Manufacturing. Nano Lett. 2021, 21, 10149–10156. [Google Scholar] [CrossRef]
- Bello-Lopez, J.M.; Silva-Bermudez, P.; Prado, G.; Martínez, A.; Ibáñez-Cervantes, G.; Cureño-Díaz, M.A.; Rocha-Zavaleta, L.; Manzo-Merino, J.; Almaguer-Flores, A.; Ramos-Vilchis, C.; et al. Biocide effect against SARS-CoV-2 and ESKAPE pathogens of a noncytotoxic silver–copper nanofilm. Biomed. Mater. 2021, 17, 015002. [Google Scholar] [CrossRef]
- Mosselhy, D.A.; Kareinen, L.; Kivistö, I.; Aaltonen, K.; Virtanen, J.; Ge, Y.; Sironen, T. Copper-Silver Nanohybrids: SARS-CoV-2 Inhibitory Surfaces. Nanomaterials 2021, 11, 1820. [Google Scholar] [CrossRef]
- Chang, S.Y.; Huang, K.Y.; Chao, T.L.; Kao, H.C.; Pang, Y.H.; Lu, L.; Chiu, C.L.; Huang, H.C.; Cheng, T.J.R.; Fang, J.M.; et al. Nanoparticle composite TPNT1 is effective against SARS-CoV-2 and influenza viruses. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Chuong, C.; DuChane, C.M.; Webb, E.M.; Rai, P.; Marano, J.M.; Bernier, C.M.; Merola, J.S.; Weger-Lucarelli, J. Noble Metal Organometallic Complexes Display Antiviral Activity against SARS-CoV-2. Viruses 2021, 13, 980. [Google Scholar] [CrossRef]
- Jaber, N.; Al-Remawi, M.; Al-Akayleh, F.; Al-Muhtaseb, N.; Al-Adham, I.S.I.; Collier, P.J. A review of the antiviral activity of Chitosan, including patented applications and its potential use against COVID-19. J. Appl. Microbiol. 2021, 132, 41–58. [Google Scholar] [CrossRef]
- Sharma, N.; Modak, C.; Singh, P.K.; Kumar, R.; Khatri, D.; Singh, S.B. Underscoring the immense potential of chitosan in fighting a wide spectrum of viruses: A plausible molecule against SARS-CoV-2? Int. J. Biol. Macromol. 2021, 179, 33–44. [Google Scholar] [CrossRef]
- Dmour, I.; Islam, N. Recent advances on chitosan as an adjuvant for vaccine delivery. Int. J. Biol. Macromol. 2022, 200, 498–519. [Google Scholar] [CrossRef]
- Safarzadeh, M.; Sadeghi, S.; Azizi, M.; Rastegari-Pouyani, M.; Pouriran, R.; Hoseini, M.H.M. Chitin and chitosan as tools to combat COVID-19: A triple approach. Int. J. Biol. Macromol. 2021, 183, 235–244. [Google Scholar] [CrossRef]
- Rahbar, M.R.; Galeh, H.E.G.; Khalili, S.; Jahangiri, A. Chitosan: A Promising Protective Component Against SARS-CoV-2 and Influenza Virus. Lett. Drug Des. Discov. 2021, 18, 418–421. [Google Scholar] [CrossRef]
- Alitongbieke, G.; Li, X.M.; Wu, Q.C.; Lin, Z.C.; Huang, J.F.; Xue, Y.; Liu, J.N.; Lin, J.M.; Pan, T.; Chen, Y.X.; et al. Effect of β-chitosan on the binding interaction between SARS-CoV-2 S-RBD and ACE2. bioRxiv 2020, 1–29. [Google Scholar] [CrossRef]
- Kalathiya, U.; Padariya, M.; Mayordomo, M.; Lisowska, M.; Nicholson, J.; Singh, A.; Baginski, M.; Fahraeus, R.; Carragher, N.; Ball, K.; et al. Highly Conserved Homotrimer Cavity Formed by the SARS-CoV-2 Spike Glycoprotein: A Novel Binding Site. J. Clin. Med. 2020, 9, 1473. [Google Scholar] [CrossRef]
- He, X.; Xing, R.; Li, K.; Qin, Y.; Zou, P.; Liu, S.; Yu, H.; Li, P. Beta-chitosan extracted from Loligo Japonica for a potential use to inhibit Newcastle disease. Int. J. Biol. Macromol. 2016, 82, 614–620. [Google Scholar] [CrossRef]
- Wang, W.; Xue, C.; Mao, X. Chitosan: Structural modification, biological activity and application. Int. J. Biol. Macromol. 2020, 164, 4532–4546. [Google Scholar] [CrossRef]
- He, X.; Xing, R.; Liu, S.; Qin, Y.; Li, K.; Yu, H.; Li, P. The improved antiviral activities of amino-modified chitosan derivatives on Newcastle virus. Drug Chem. Toxicol. 2019, 44, 335–340. [Google Scholar] [CrossRef]
- Artan, M.; Karadeniz, F.; Karagozlu, M.Z.; Kim, M.M.; Kim, S.K. Anti-HIV-1 activity of low molecular weight sulfated chitooligosaccharides. Carbohydr. Res. 2010, 345, 656–662. [Google Scholar] [CrossRef]
- Milewska, A.; Kaminski, K.; Ciejka, J.; Kosowicz, K.; Zeglen, S.; Wojarski, J.; Nowakowska, M.; Szczubiałka, K.; Pyrc, K. HTCC: Broad Range Inhibitor of Coronavirus Entry. PLoS ONE 2016, 11, e0156552. [Google Scholar] [CrossRef] [PubMed]
- Milewska, A.; Chi, Y.; Szczepanski, A.; Barreto-Duran, E.; Dabrowska, A.; Botwina, P.; Obloza, M.; Liu, K.; Liu, D.; Guo, X.; et al. HTCC as a Polymeric Inhibitor of SARS-CoV-2 and MERS-CoV. J. Virol. 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Milewska, A.; Ciejka, J.; Kaminski, K.; Karewicz, A.; Bielska, D.; Zeglen, S.; Karolak, W.; Nowakowska, M.; Potempa, J.; Bosch, B.J.; et al. Novel polymeric inhibitors of HCoV-NL63. Antivir. Res. 2013, 97, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Pyrć, K.; Milewska, A.; Duran, E.B.; Botwina, P.; Dabrowska, A.; Jedrysik, M.; Benedyk, M.; Lopes, R.; Arenas-Pinto, A.; Badr, M.; et al. SARS-CoV-2 inhibition using a mucoadhesive, amphiphilic chitosan that may serve as an anti-viral nasal spray. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Vörös-Horváth, B.; Živković, P.; Bánfai, K.; Bóvári-Biri, J.; Pongrácz, J.; Bálint, G.; Pál, S.; Széchenyi, A. Preparation and Characterization of ACE2 Receptor Inhibitor-Loaded Chitosan Hydrogels for Nasal Formulation to Reduce the Risk of COVID-19 Viral Infection. ACS Omega 2022, 7, 3240–3253. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, N.A.; El-Kemary, M.A. Silymarin/curcumin loaded albumin nanoparticles coated by chitosan as muco-inhalable delivery system observing anti-inflammatory and anti COVID-19 characterizations in oleic acid triggered lung injury and in vitro COVID-19 experiment. Int. J. Biol. Macromol. 2022, 198, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lu, C.; Ding, Y.; Zhang, J.; Tan, F.; Liu, J.; Yang, G.; Wang, Y.; Li, Z.; Yang, M.; et al. Red blood cell-hitchhiking mediated pulmonary delivery of ivermectin: Effects of nanoparticle properties. Int. J. Pharm. 2022, 619, 121719. [Google Scholar] [CrossRef]
- Brenner, J.S.; Mitragotri, S.; Muzykantov, V.R. Red Blood Cell Hitchhiking: A Novel Approach for Vascular Delivery of Nanocarriers. Annu. Rev. Biomed. Eng. 2021, 23, 225–248. [Google Scholar] [CrossRef]
- Qingliang, Y.; Yang, C.; Wang, R.; Xiao, B.; Jiang, Z.; Xu, H.; Wang, J. Chitosan Hydrogel, Preparation Method Thereof, Antiviral Spray and Antiviral Liquid Glove. Chinese Patent Publication no. 111529762, 2020. Available online: https://patents.google.com/patent/CN111529762A/en?oq=111529762 (accessed on 15 June 2022).
- Zhang, S.; Zhang, Q.; Chen, J.; Dong, H.; Cui, A.; Sun, L.; Wang, N.; Li, J.; Qu, Z. Cost-effective chitosan thermal bonded nonwovens serving as an anti-viral inhibitor layer in face mask. Mater. Lett. 2022, 318, 132203. [Google Scholar] [CrossRef]
- Xianming, S.; Qi, Y.; Li, J.; Hao, Y.; Zhang, D. Antiviral Filter Layer Prepared from Copper-Containing Chitosan Fiber and Application Thereof. Chinese Patent Publication no. 111235871, 2020. Available online: https://patents.google.com/patent/CN111235871A/en?oq=111235871 (accessed on 15 June 2022).
- Favatela, M.F.; Otarola, J.; Ayala-Peña, V.B.; Dolcini, G.; Perez, S.; Nicolini, A.T.; Alvarez, V.A.; Lassalle, V.L. Development and Characterization of Antimicrobial Textiles from Chitosan-Based Compounds: Possible Biomaterials Against SARS-CoV-2 Viruses. J. Inorg. Organomet. Polym. Mater. 2022, 32, 1473–1486. [Google Scholar] [CrossRef]
- Rajakumar, G.; Zhang, X.H.; Gomathi, T.; Wang, S.F.; Ansari, M.A.; Mydhili, G.; Nirmala, G.; Alzohairy, M.A.; Chung, I.M. Current Use of Carbon-Based Materials for Biomedical Applications—A Prospective and Review. Processes 2020, 8, 355. [Google Scholar] [CrossRef]
- Caccamo, D.; Currò, M.; Ientile, R.; Verderio, E.A.; Scala, A.; Mazzaglia, A.; Pennisi, R.; Musarra-Pizzo, M.; Zagami, R.; Neri, J.; et al. Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration. Int. J. Mol. Sci. 2020, 21, 4891. [Google Scholar] [CrossRef]
- Nimushakavi, S.; Haque, S.; Kotcherlakota, R.; Patra, C.R. Biomedical Applications of Carbon Nanotubes: Recent Development and Future Challenges. In Nanoengineering of Biomaterials; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2022; Chapter 12; pp. 353–388. [Google Scholar] [CrossRef]
- Serrano-Aroca, Á.; Takayama, K.; Tuñón-Molina, A.; Seyran, M.; Hassan, S.S.; Choudhury, P.P.; Uversky, V.N.; Lundstrom, K.; Adadi, P.; Palù, G.; et al. Carbon-Based Nanomaterials: Promising Antiviral Agents to Combat COVID-19 in the Microbial-Resistant Era. ACS Nano 2021, 15, 8069–8086. [Google Scholar] [CrossRef]
- d’Amora, M.; Giordani, S. Carbon Nanomaterials for Nanomedicine. In Smart Nanoparticles for Biomedicine; Elsevier: Amsterdam, The Netherlands, 2018; pp. 103–113. [Google Scholar] [CrossRef]
- Piperno, A.; Scala, A.; Mazzaglia, A.; Neri, J.; Pennisi, R.; Sciortino, M.; Grassi, G. Cellular Signaling Pathways Activated by Functional Graphene Nanomaterials. Int. J. Mol. Sci. 2018, 19, 3365. [Google Scholar] [CrossRef]
- Skariyachan, S.; Gopal, D.; Deshpande, D.; Joshi, A.; Uttarkar, A.; Niranjan, V. Carbon fullerene and nanotube are probable binders to multiple targets of SARS-CoV-2: Insights from computational modeling and molecular dynamic simulation studies. Infect. Genet. Evol. 2021, 96, 105155. [Google Scholar] [CrossRef]
- Jomhori, M.; Mosaddeghi, H.; Farzin, H. Tracking the interaction between single-wall carbon nanotube and SARS-Cov-2 spike glycoprotein: A molecular dynamics simulations study. Comput. Biol. Med. 2021, 136, 104692. [Google Scholar] [CrossRef]
- Suárez, M.; Makowski, K.; Lemos, R.; Almagro, L.; Rodríguez, H.; Herranz, M.Á.; Molero, D.; Ortiz, O.; Maroto, E.; Albericio, F.; et al. An Androsterone-H2@C60 hybrid: Synthesis, Properties and Molecular Docking Simulations with SARS-Cov-2. ChemPlusChem 2021, 86, 972–981. [Google Scholar] [CrossRef]
- Tomo, S.; Banerjee, M.; Sharma, P.; Garg, M. Does dehydroepiandrosterone sulfate have a role in COVID-19 prognosis and treatment? Endocr. Regul. 2021, 55, 174–181. [Google Scholar] [CrossRef]
- Unal, M.A.; Bayrakdar, F.; Nazir, H.; Besbinar, O.; Gurcan, C.; Lozano, N.; Arellano, L.M.; Yalcin, S.; Panatli, O.; Celik, D.; et al. Graphene Oxide Nanosheets Interact and Interfere with SARS-CoV-2 Surface Proteins and Cell Receptors to Inhibit Infectivity. Small 2021, 17, 2101483. [Google Scholar] [CrossRef]
- Maio, F.D.; Palmieri, V.; Babini, G.; Augello, A.; Palucci, I.; Perini, G.; Salustri, A.; Spilman, P.; Spirito, M.D.; Sanguinetti, M.; et al. Graphene nanoplatelet and graphene oxide functionalization of face mask materials inhibits infectivity of trapped SARS-CoV-2. iScience 2021, 24, 102788. [Google Scholar] [CrossRef]
- Stagi, L.; Forni, D.D.; Malfatti, L.; Caboi, F.; Salis, A.; Poddesu, B.; Cugia, J.; Lori, F.; Galleri, G.; Innocenzi, P. Effective SARS-CoV-2 antiviral activity of hyperbranched polylysine nanopolymers. Nanoscale 2021, 13, 16465–16476. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.H.; Choi, S.H.; Smith, S.A. Polyphosphate: An ancient molecule that links platelets, coagulation, and inflammation. Blood 2012, 119, 5972–5979. [Google Scholar] [CrossRef] [PubMed]
- Neufurth, M.; Wang, X.; Tolba, E.; Lieberwirth, I.; Wang, S.; Schröder, H.C.; Müller, W.E. The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem. Pharmacol. 2020, 182, 114215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, W.; Jin, Q.; Pan, F.; Zhu, J.; Liu, Y.; Chen, L.; Shen, J.; Yang, Y.; Chen, Q.; et al. Inhalable nanocatchers for SARS-CoV-2 inhibition. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Salazar-García, M.; Acosta-Contreras, S.; Rodríguez-Martínez, G.; Cruz-Rangel, A.; Flores-Alanis, A.; Patiño-López, G.; Luna-Pineda, V.M. Pseudotyped Vesicular Stomatitis Virus-Severe Acute Respiratory Syndrome-Coronavirus-2 Spike for the Study of Variants, Vaccines, and Therapeutics Against Coronavirus Disease 2019. Front. Microbiol. 2022, 12, 817200. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Chen, Y.; Zhao, J.; Han, S.; Zhao, G.; Kang, J.; Liu, Y.; Wang, L.; Wang, X.; et al. Membrane Nanoparticles Derived from ACE2-Rich Cells Block SARS-CoV-2 Infection. ACS Nano 2021, 15, 6340–6351. [Google Scholar] [CrossRef]
- Zhang, Q.; Honko, A.; Zhou, J.; Gong, H.; Downs, S.N.; Vasquez, J.H.; Fang, R.H.; Gao, W.; Griffiths, A.; Zhang, L. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nano Lett. 2020, 20, 5570–5574. [Google Scholar] [CrossRef]
- Rao, L.; Xia, S.; Xu, W.; Tian, R.; Yu, G.; Gu, C.; Pan, P.; Meng, Q.F.; Cai, X.; Qu, D.; et al. Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2020, 117, 27141–27147. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Z.; Dinh, P.U.C.; Zhu, D.; Popowski, K.D.; Lutz, H.; Hu, S.; Lewis, M.G.; Cook, A.; Andersen, H.; et al. Cell-mimicking nanodecoys neutralize SARS-CoV-2 and mitigate lung injury in a non-human primate model of COVID-19. Nat. Nanotechnol. 2021, 16, 942–951. [Google Scholar] [CrossRef]
- Chen, M.; Rosenberg, J.; Cai, X.; Lee, A.C.H.; Shi, J.; Nguyen, M.; Wignakumar, T.; Mirle, V.; Edobor, A.J.; Fung, J.; et al. Nanotraps for the containment and clearance of SARS-CoV-2. Matter 2021, 4, 2059–2082. [Google Scholar] [CrossRef]
- Wu, Y.; Tibrewal, N.; Birge, R.B. Phosphatidylserine recognition by phagocytes: A view to a kill. Trends Cell Biol. 2006, 16, 189–197. [Google Scholar] [CrossRef]
- Shah, N.K.; Gupta, S.K.; Wang, Z.; Meenach, S.A. Enhancement of macrophage uptake via phosphatidylserine-coated acetalated dextran nanoparticles. J. Drug Deliv. Sci. Technol. 2019, 50, 57–65. [Google Scholar] [CrossRef]
- Strong, T.A.; Pelaez, D. ACE2-cytomimetic particles restrict SARS-Cov-2 spike protein binding to cellular targets. Biotechnol. Rep. 2021, 32, e00681. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Ameratunga, R.; Lehnert, K.; Leung, E.; Comoletti, D.; Snell, R.; Woon, S.T.; Abbott, W.; Mears, E.; Steele, R.; McKee, J.; et al. Inhaled modified angiotensin converting enzyme 2 (ACE2) as a decoy to mitigate SARS-CoV-2 infection. N. Z. Med. J. 2020, 133, 112–118. [Google Scholar]
- Kim, J.; Mukherjee, A.; Nelson, D.; Jozic, A.; Sahay, G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. bioRxiv 2020, 1–44. [Google Scholar] [CrossRef]
- Cai, X.; Chen, M.; Prominski, A.; Lin, Y.; Ankenbruck, N.; Rosenberg, J.; Nguyen, M.; Shi, J.; Tomatsidou, A.; Randall, G.; et al. A Multifunctional Neutralizing Antibody-Conjugated Nanoparticle Inhibits and Inactivates SARS-CoV-2. Adv. Sci. 2021, 9, 2103240. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, C.; Wu, W.; Wang, F.; Du, L.; Zhang, X.; Xiong, Y.; He, X.; Cai, Y.; Kwok, R.T.K.; et al. Highly efficient photothermal nanoagent achieved by harvesting energy via excited-state intramolecular motion within nanoparticles. Nat. Commun. 2019, 10, 768. [Google Scholar] [CrossRef]
- Demidova, T.N.; Hamblin, M.R. Photodynamic Therapy Targeted to Pathogens. Int. J. Immunopathol. Pharmacol. 2004, 17, 245–254. [Google Scholar] [CrossRef]
- Wiehe, A.; O’Brien, J.M.; Senge, M.O. Trends and targets in antiviral phototherapy. Photochem. Photobiol. Sci. 2019, 18, 2565–2612. [Google Scholar] [CrossRef]
- Pourhajibagher, M.; Azimi, M.; Haddadi-Asl, V.; Ahmadi, H.; Gholamzad, M.; Ghorbanpour, S.; Bahador, A. Robust antimicrobial photodynamic therapy with curcumin-poly (lactic-co-glycolic acid) nanoparticles against COVID-19: A preliminary in vitro study in Vero cell line as a model. Photodiagnosis Photodyn. Ther. 2021, 34, 102286. [Google Scholar] [CrossRef] [PubMed]
- Panhóca, V.H.; Florez, F.L.E.; Corrêa, T.Q.; Paolillo, F.R.; de Souza, C.W.O.; Bagnato, V.S. Oral Decontamination of Orthodontic Patients Using Photodynamic Therapy Mediated by Blue-Light Irradiation and Curcumin Associated with Sodium Dodecyl Sulfate. Photomed. Laser Surg. 2016, 34, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Khaiboullina, S.; Uppal, T.; Dhabarde, N.; Subramanian, V.R.; Verma, S.C. Inactivation of Human Coronavirus by Titania Nanoparticle Coatings and UVC Radiation: Throwing Light on SARS-CoV-2. Viruses 2020, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, J.; Zarzyńska, J.; Pławińska-Czarnak, J. Comparison of Infectious Agents Susceptibility to Photocatalytic Effects of Nanosized Titanium and Zinc Oxides: A Practical Approach. Nanoscale Res. Lett. 2015, 10, 309. [Google Scholar] [CrossRef]
- Norrrahim, M.N.F.; Kasim, N.A.M.; Knight, V.F.; Ong, K.K.; Noor, S.A.M.; Jamal, S.H.; Shah, N.A.A.; Halim, N.A.; Ilyas, R.A.; Yunus, W.M.Z.W. Cationic Nanocellulose as Promising Candidate for Filtration Material of COVID-19: A Perspective. Appl. Sci. Eng. Prog. 2021, 14, 580–587. [Google Scholar] [CrossRef]
- Gunathilake, T.M.S.U.; Ching, Y.C.; Chuah, C.H.; Rahman, N.A.; Liou, N.S. Recent advances in celluloses and their hybrids for stimuli-responsive drug delivery. Int. J. Biol. Macromol. 2020, 158, 670–688. [Google Scholar] [CrossRef]
- Gunathilake, T.M.S.U.; Ching, Y.C.; Uyama, H.; Hai, N.D.; Chuah, C.H. Enhanced curcumin loaded nanocellulose: A possible inhalable nanotherapeutic to treat COVID-19. Cellulose 2022, 29, 1821–1840. [Google Scholar] [CrossRef]
- Fischer, H.; Widdicombe, J.H. Mechanisms of Acid and Base Secretion by the Airway Epithelium. J. Membr. Biol. 2006, 211, 139–150. [Google Scholar] [CrossRef]
- Sviridov, D.; Miller, Y.I.; Ballout, R.A.; Remaley, A.T.; Bukrinsky, M. Targeting Lipid Rafts—A Potential Therapy for COVID-19. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Paolacci, S.; Kiani, A.K.; Shree, P.; Tripathi, D.; Tripathi, Y.B.; Tripathi, P.; Tartaglia, G.M.; Farronato, M.; Farronato, G.; Connelly, S.T.; et al. Scoping review on the role and interactions of hydroxytyrosol and alpha-cyclodextrin in lipid-raft-mediated endocytosis of SARS-CoV-2 and bioinformatic molecular docking studies. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 90–100. [Google Scholar] [CrossRef]
- Kiani, A.K.; Dhuli, K.; Anpilogov, K.; Bressan, S.; Dautaj, A.; Dundar, M.; Beccari, T.; Ergoren, M.C.; Bertelli, M. Natural compounds as inhibitors of SARS-CoV-2 endocytosis: A promising approach against COVID-19. Acta Bio. Med. Atenei Parm. 2020, 91, e2020008. [Google Scholar] [CrossRef]
- Monteiro, M.; Silva, A.F.R.; Resende, D.; Braga, S.S.; Coimbra, M.A.; Silva, A.M.S.; Cardoso, S.M. Strategies to Broaden the Applications of Olive Biophenols Oleuropein and Hydroxytyrosol in Food Products. Antioxidants 2021, 10, 444. [Google Scholar] [CrossRef]
- Anwer, M.K.; Ali, E.A.; Iqbal, M.; Ahmed, M.M.; Aldawsari, M.F.; Saqr, A.A.; Ansari, M.N.; Aboudzadeh, M.A. Development of Sustained Release Baricitinib Loaded Lipid-Polymer Hybrid Nanoparticles with Improved Oral Bioavailability. Molecules 2021, 27, 168. [Google Scholar] [CrossRef]
- Khater, S.E.; El-khouly, A.; Abdel-Bar, H.M.; Al-mahallawi, A.M.; Ghorab, D.M. Fluoxetine hydrochloride loaded lipid polymer hybrid nanoparticles showed possible efficiency against SARS-CoV-2 infection. Int. J. Pharm. 2021, 607, 121023. [Google Scholar] [CrossRef]
- Dechaumes, A.; Nekoua, M.P.; Belouzard, S.; Sane, F.; Engelmann, I.; Dubuisson, J.; Alidjinou, E.K.; Hober, D. Fluoxetine Can Inhibit SARS-CoV-2 In Vitro. Microorganisms 2021, 9, 339. [Google Scholar] [CrossRef]
- Hoertel, N.; Sánchez-Rico, M.; Vernet, R.; Beeker, N.; Jannot, A.S.; Neuraz, A.; Salamanca, E.; Paris, N.; Daniel, C.; Gramfort, A.; et al. Association between antidepressant use and reduced risk of intubation or death in hospitalized patients with COVID-19: Results from an observational study. Mol. Psychiatry 2021, 26, 5199–5212. [Google Scholar] [CrossRef]
- Jermy, B.R.; Ravinayagam, V.; Almohazey, D.; Alamoudi, W.; Dafalla, H.; Akhtar, S.; Tanimu, G. PEGylated green halloysite/spinel ferrite nanocomposites for pH sensitive delivery of dexamethasone: A potential pulmonary drug delivery treatment option for COVID-19. Appl. Clay Sci. 2022, 216, 106333. [Google Scholar] [CrossRef]
- Dong, J.; Cheng, Z.; Tan, S.; Zhu, Q. Clay nanoparticles as pharmaceutical carriers in drug delivery systems. Expert Opin. Drug Deliv. 2020, 18, 695–714. [Google Scholar] [CrossRef]
- Amiri, M.; Salavati-Niasari, M.; Akbari, A. Magnetic nanocarriers: Evolution of spinel ferrites for medical applications. Adv. Colloid Interface Sci. 2019, 265, 29–44. [Google Scholar] [CrossRef]
- Pindiprolu, S.K.S.; Kumar, C.S.P.; Golla, V.S.K.; Likitha, P.; Chandra, S.; Esub Basha, S.K.; Ramachandra, R.K. Pulmonary delivery of nanostructured lipid carriers for effective repurposing of salinomycin as an antiviral agent. Med. Hypotheses 2020, 143, 109858. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Fatima, W.; Alzahrani, A.K.; Suhail, N.; Alshammari, M.K.; Alghitran, A.A.; Alshammari, F.N.; Ghoneim, M.M.; Alshehri, S.; Shakeel, F. Development of Therapeutic and Prophylactic Zinc Compositions for Use against COVID-19: A Glimpse of the Trends, Inventions, and Patents. Nutrients 2022, 14, 1227. [Google Scholar] [CrossRef] [PubMed]
- Balboni, E.; Zagnoli, F.; Filippini, T.; Fairweather-Tait, S.J.; Vinceti, M. Zinc and selenium supplementation in COVID-19 prevention and treatment: A systematic review of the experimental studies. J. Trace Elem. Med. Biol. 2022, 71, 126956. [Google Scholar] [CrossRef] [PubMed]
- Ghareeb, D.A.; Saleh, S.R.; Seadawy, M.G.; Nofal, M.S.; Abdulmalek, S.A.; Hassan, S.F.; Khedr, S.M.; AbdElwahab, M.G.; Sobhy, A.A.; saber Ali Abdel-Hamid, A.; et al. Nanoparticles of ZnO/Berberine complex contract COVID-19 and respiratory co-bacterial infection in addition to elimination of hydroxychloroquine toxicity. J. Pharm. Investig. 2021, 51, 735–757. [Google Scholar] [CrossRef]
- Warowicka, A.; Nawrot, R.; Goździcka-Józefiak, A. Antiviral activity of berberine. Arch. Virol. 2020, 165, 1935–1945. [Google Scholar] [CrossRef]
- AbouAitah, K.; Allayh, A.K.; Wojnarowicz, J.; Shaker, Y.M.; Swiderska-Sroda, A.; Lojkowski, W. Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses. Pharmaceutics 2021, 13, 2174. [Google Scholar] [CrossRef]
- Chang, C.W.; Lee, M.C.; Lin, B.R.; Lu, Y.P.; Hsu, Y.J.; Chuang, C.Y.; Huang, T.T.; Chen, Y.K. Azithromycin Plus Zinc Sulfate Rapidly and Synergistically Suppresses IκBα-Mediated In Vitro Human Airway Cell ACE2 Expression for SARS-CoV-2 Entry. bioRxiv 2021, 1–23. [Google Scholar] [CrossRef]
- Wu, J.; Wang, H.; Li, B. Structure-aided ACEI-capped remdesivir-loaded novel PLGA nanoparticles: Toward a computational simulation design for anti-SARS-CoV-2 therapy. Phys. Chem. Chem. Phys. 2020, 22, 28434–28439. [Google Scholar] [CrossRef]
- Vartak, R.; Patil, S.M.; Saraswat, A.; Patki, M.; Kunda, N.K.; Patel, K. Aerosolized nanoliposomal carrier of remdesivir: An effective alternative for COVID-19 treatment in vitro. Nanomedicine 2021, 16, 1187–1202. [Google Scholar] [CrossRef]
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.; Soemardy, C.; et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226. [Google Scholar] [CrossRef]
- Wu, S.Y.; Putral, L.N.; Liang, M.; Chang, H.I.; Davies, N.M.; McMillan, N.A.J. Development of a Novel Method for Formulating Stable siRNA-Loaded Lipid Particles for In vivo Use. Pharm. Res. 2008, 26, 512–522. [Google Scholar] [CrossRef]
- Surnar, B.; Shah, A.S.; Guin, S.; Kolishetti, N.; Fornoni, A.; Dhar, S. Blending of Designer Synthetic Polymers to a Dual Targeted Nanoformulation for SARS-CoV-2 Associated Kidney Damage. Biomacromolecules 2021, 22, 4244–4250. [Google Scholar] [CrossRef]
- Yoshida, M.; Claypool, S.M.; Wagner, J.S.; Mizoguchi, E.; Mizoguchi, A.; Roopenian, D.C.; Lencer, W.I.; Blumberg, R.S. Human Neonatal Fc Receptor Mediates Transport of IgG into Luminal Secretions for Delivery of Antigens to Mucosal Dendritic Cells. Immunity 2004, 20, 769–783. [Google Scholar] [CrossRef]
- Rhee, H.; Ng, K.L.; Tse, B.W.C.; Yeh, M.C.; Russell, P.J.; Nelson, C.; Thomas, P.; Samaratunga, H.; Vela, I.; Gobe, G.; et al. Using prostate specific membrane antigen (PSMA) expression in clear cell renal cell carcinoma for imaging advanced disease. Pathology 2016, 48, 613–616. [Google Scholar] [CrossRef]
- Giofrè, S.V.; Napoli, E.; Iraci, N.; Speciale, A.; Cimino, F.; Muscarà, C.; Molonia, M.S.; Ruberto, J.; Saija, A. Interaction of selected terpenoids with two SARS-CoV-2 key therapeutic targets: An in silico study through molecular docking and dynamics simulations. Comput. Biol. Med. 2021, 134, 104538. [Google Scholar] [CrossRef]
- Ha, D.P.; Krieken, R.V.; Carlos, A.J.; Lee, A.S. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 2020, 81, 452–482. [Google Scholar] [CrossRef]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Lee, A.S. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 2005, 35, 373–381. [Google Scholar] [CrossRef]
- Li, J.; Lee, A. Stress Induction of GRP78/BiP and Its Role in Cancer. Curr. Mol. Med. 2006, 6, 45–54. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, R.; Jung, D.Y.; Barron, E.; Friedline, R.H.; Benoit, V.M.; Hinton, D.R.; Kim, J.K.; Lee, A.S. GRP78 plays an essential role in adipogenesis and postnatal growth in mice. FASEB J. 2012, 27, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Toyoda, S.; Nishitani, S.; Fukuhara, A.; Kita, S.; Otsuki, M.; Shimomura, I. Possible Involvement of Adipose Tissue in Patients With Older Age, Obesity, and Diabetes With SARS-CoV-2 Infection (COVID-19) via GRP78 (BIP/HSPA5): Significance of Hyperinsulinemia Management in COVID-19. Diabetes 2021, 70, 2745–2755. [Google Scholar] [CrossRef] [PubMed]
- Gammone, M.A.; D’Orazio, N. COVID-19 and Obesity: Overlapping of Two Pandemics. Obes. Facts 2021, 14, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Dugail, I.; Amri, E.Z.; Vitale, N. High prevalence for obesity in severe COVID-19: Possible links and perspectives towards patient stratification. Biochimie 2020, 179, 257–265. [Google Scholar] [CrossRef]
- Guo, W.; Wong, S.; Xie, W.; Lei, T.; Luo, Z. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3-L1 and rat primary preadipocytes. Am. J. Physiol.-Endocrinol. Metab. 2007, 293, E576–E586. [Google Scholar] [CrossRef]
- Perveen, S. Introductory Chapter: Terpenes and Terpenoids. In Terpenes and Terpenoids; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Boncan, D.A.T.; Tsang, S.S.; Li, C.; Lee, I.H.; Lam, H.M.; Chan, T.F.; Hui, J.H. Terpenes and Terpenoids in Plants: Interactions with Environment and Insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Tariq, S.; Wani, S.; Rasool, W.; Shafi, K.; Bhat, M.A.; Prabhakar, A.; Shalla, A.H.; Rather, M.A. A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens. Microb. Pathog. 2019, 134, 103580. [Google Scholar] [CrossRef]
- Prado-Audelo, M.L.D.; Cortés, H.; Caballero-Florán, I.H.; González-Torres, M.; Escutia-Guadarrama, L.; Bernal-Chávez, S.A.; Giraldo-Gomez, D.M.; Magaña, J.J.; Leyva-Gómez, G. Therapeutic Applications of Terpenes on Inflammatory Diseases. Front. Pharmacol. 2021, 12, 2114. [Google Scholar] [CrossRef]
- Kamran, S.; Sinniah, A.; Abdulghani, M.A.M.; Alshawsh, M.A. Therapeutic Potential of Certain Terpenoids as Anticancer Agents: A Scoping Review. Cancers 2022, 14, 1100. [Google Scholar] [CrossRef]
- Duschatzky, C.B.; Possetto, M.L.; Talarico, L.B.; García, C.C.; Michis, F.; Almeida, N.V.; de Lampasona, M.P.; Schuff, C.; Damonte, E.B. Evaluation of Chemical and Antiviral Properties of Essential Oils from South American Plants. Antivir. Chem. Chemother. 2005, 16, 247–251. [Google Scholar] [CrossRef]
- Loizzo, M.; Saab, A.; Tundis, R.; Statti, G.; Menichini, F.; Lampronti, I.; Gambari, R.; Cinatl, J.; Doerr, H. Phytochemical Analysis and in vitro Antiviral Activities of the Essential Oils of Seven Lebanon Species. Chem. Biodivers. 2008, 5, 461–470. [Google Scholar] [CrossRef]
- Astani, A.; Reichling, J.; Schnitzler, P. Comparative study on the antiviral activity of selected monoterpenes derived from essential oils. Phytother. Res. 2009, 24, 673–679. [Google Scholar] [CrossRef]
- Armaka, M.; Papanikolaou, E.; Sivropoulou, A.; Arsenakis, M. Antiviral properties of isoborneol, a potent inhibitor of herpes simplex virus type 1. Antivir. Res. 1999, 43, 79–92. [Google Scholar] [CrossRef]
- Kurokawa, M.; Basnet, P.; Ohsugi, M.; Hozumi, T.; Kadota, S.; Namba, T.; Kawana, T.; Shiraki, K. Anti-Herpes Simplex Virus Activity of Moronic Acid Purified from Rhus javanica In Vitro and In Vivo. J. Pharmacol. Exp. Ther. 1999, 289, 72–78. [Google Scholar]
- Brezáni, V.; Leláková, V.; Hassan, S.; Berchová-Bímová, K.; Nový, P.; Klouček, P.; Maršík, P.; Dall’Acqua, S.; Hošek, J.; Šmejkal, K. Anti-Infectivity against Herpes Simplex Virus and Selected Microbes and Anti-Inflammatory Activities of Compounds Isolated from Eucalyptus globulus Labill. Viruses 2018, 10, 360. [Google Scholar] [CrossRef]
- Lin, L.T.; Chung, C.Y.; Hsu, W.C.; Chang, S.P.; Hung, T.C.; Shields, J.; Russell, R.S.; Lin, C.C.; Li, C.F.; Yen, M.H.; et al. Saikosaponin b2 is a naturally occurring terpenoid that efficiently inhibits hepatitis C virus entry. J. Hepatol. 2015, 62, 541–548. [Google Scholar] [CrossRef]
- Chung, C.Y.; Liu, C.H.; Burnouf, T.; Wang, G.H.; Chang, S.P.; Jassey, A.; Tai, C.J.; Tai, C.J.; Huang, C.J.; Richardson, C.D.; et al. Activity-based and fraction-guided analysis of Phyllanthus urinaria identifies loliolide as a potent inhibitor of hepatitis C virus entry. Antivir. Res. 2016, 130, 58–68. [Google Scholar] [CrossRef]
- Wohlfarth, C.; Efferth, T. Natural products as promising drug candidates for the treatment of hepatitis B and C. Acta Pharmacol. Sin. 2008, 30, 25–30. [Google Scholar] [CrossRef]
- Huan, C.; Xu, Y.; Zhang, W.; Guo, T.; Pan, H.; Gao, S. Research Progress on the Antiviral Activity of Glycyrrhizin and its Derivatives in Liquorice. Front. Pharmacol. 2021, 12, 680674. [Google Scholar] [CrossRef]
- Fujioka, T.; Kashiwada, Y.; Kilkuskie, R.E.; Cosentino, L.M.; Ballas, L.M.; Jiang, J.B.; Janzen, W.P.; Chen, I.S.; Lee, K.H. Anti-AIDS Agents, 11. Betulinic Acid and Platanic Acid as Anti-HIV Principles from SyziJunm claviflorum, and the Anti-HIV Activity of Structurally Related Triterpenoids. J. Nat. Prod. 1994, 57, 243–247. [Google Scholar] [CrossRef]
- Cichewicz, R.H.; Kouzi, S.A. Chemistry, biological activity, and chemotherapeutic potential of betulinic acid for the prevention and treatment of cancer and HIV infection. Med. Res. Rev. 2003, 24, 90–114. [Google Scholar] [CrossRef] [PubMed]
- Medina-O’Donnell, M.; Rivas, F.; Reyes-Zurita, F.J.; Cano-Muñoz, M.; Martinez, A.; Lupiañez, J.A.; Parra, A. Oleanolic Acid Derivatives as Potential Inhibitors of HIV-1 Protease. J. Nat. Prod. 2019, 82, 2886–2896. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, J.H.; Kim, Y.M.; Jeong, H.J.; Kim, D.W.; Park, K.H.; Kwon, H.J.; Park, S.J.; Lee, W.S.; Ryu, Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorganic Med. Chem. 2012, 20, 5928–5935. [Google Scholar] [CrossRef]
- Chang, F.R.; Yen, C.T.; EI-Shazly, M.; Lin, W.H.; Yen, M.H.; Lin, K.H.; Wu, Y.C. Anti-Human Coronavirus (anti-HCoV) Triterpenoids from the Leaves of Euphorbia neriifolia. Nat. Prod. Commun. 2012, 7, 1934578X1200701. [Google Scholar] [CrossRef]
- Ryu, Y.B.; Park, S.J.; Kim, Y.M.; Lee, J.Y.; Seo, W.D.; Chang, J.S.; Park, K.H.; Rho, M.C.; Lee, W.S. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorganic Med. Chem. Lett. 2010, 20, 1873–1876. [Google Scholar] [CrossRef]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef]
- Yu, S.; Zhu, Y.; Xu, J.; Yao, G.; Zhang, P.; Wang, M.; Zhao, Y.; Lin, G.; Chen, H.; Chen, L.; et al. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine 2021, 85, 153364. [Google Scholar] [CrossRef]
- Li, J.; Xu, D.; Wang, L.; Zhang, M.; Zhang, G.; Li, E.; He, S. Glycyrrhizic Acid Inhibits SARS-CoV-2 Infection by Blocking Spike Protein-Mediated Cell Attachment. Molecules 2021, 26, 6090. [Google Scholar] [CrossRef]
- Ahmad, S.; Waheed, Y.; Abro, A.; Abbasi, S.W.; Ismail, S. Molecular screening of glycyrrhizin-based inhibitors against ACE2 host receptor of SARS-CoV-2. J. Mol. Model. 2021, 27, 206. [Google Scholar] [CrossRef]
- Murck, H. Symptomatic Protective Action of Glycyrrhizin (Licorice) in COVID-19 Infection? Front. Immunol. 2020, 11, 1239. [Google Scholar] [CrossRef]
- Luo, P.; Liu, D.; Li, J. Pharmacological perspective: Glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105995. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y. Potential interventions for novel coronavirus in China: A systematic review. J. Med Virol. 2020, 92, 479–490. [Google Scholar] [CrossRef]
- Gomaa, A.A.; Abdel-Wadood, Y.A. The potential of glycyrrhizin and licorice extract in combating COVID-19 and associated conditions. Phytomedicine Plus 2021, 1, 100043. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Santos, K.B.; Guedes, I.A.; Karl, A.L.M.; Dardenne, L.E. Highly Flexible Ligand Docking: Benchmarking of the DockThor Program on the LEADS-PEP Protein–Peptide Data Set. J. Chem. Inf. Model. 2020, 60, 667–683. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Kalhor, H.; Sadeghi, S.; Abolhasani, H.; Kalhor, R.; Rahimi, H. Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2S protein and human ACE2 interaction through virtual screening approaches. J. Biomol. Struct. Dyn. 2020, 40, 1299–1315. [Google Scholar] [CrossRef]
- Emon, N.U.; Alam, M.M.; Akter, I.; Akhter, S.; Sneha, A.A.; Irtiza, M.; Afroj, M.; Munni, A.; Chowdhury, M.H.; Hossain, S. Virtual screenings of the bioactive constituents of tea, prickly chaff, catechu, lemon, black pepper, and synthetic compounds with the main protease (Mpro) and human angiotensin-converting enzyme 2 (ACE 2) of SARS-CoV-2. Future J. Pharm. Sci. 2021, 7, 121. [Google Scholar] [CrossRef]
- Gyebi, G.A.; Ogunyemi, O.M.; Ibrahim, I.M.; Ogunro, O.B.; Adegunloye, A.P.; Afolabi, S.O. SARS-CoV-2 host cell entry: An in silico investigation of potential inhibitory roles of terpenoids. J. Genet. Eng. Biotechnol. 2021, 19. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Mangal, M.; Sagar, P.; Singh, H.; Raghava, G.P.S.; Agarwal, S.M. NPACT: Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target database. Nucleic Acids Res. 2012, 41, D1124–D1129. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, A.; Ashfaq, U.A.; ul Qamar, M.T.; Anwar, F.; Gulzar, F.; Ali, M.A.; Saari, N.; Pervez, M.T. MPD3: A useful medicinal plants database for drug designing. Nat. Prod. Res. 2016, 31, 1228–1236. [Google Scholar] [CrossRef]
- Muhseen, Z.T.; Hameed, A.R.; Al-Hasani, H.M.; ul Qamar, M.T.; Li, G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: Integrated computational approach. J. Mol. Liq. 2020, 320, 114493. [Google Scholar] [CrossRef] [PubMed]
- Spoel, D.V.D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Sacerdoti, F.D.; Salmon, J.K.; Shan, Y.; Shaw, D.E.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; et al. Molecular dynamics—Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing—SC’06, Tampa, FL, USA, 11–17 November 2006. [Google Scholar] [CrossRef]
- Aminu, S.; Ibrahim, M.A.; Sallau, A.B. Interaction of SARS-CoV-2 spike protein with angiotensin converting enzyme inhibitors and selected compounds from the chemical entities of biological interest. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.A.; Nagarajan, S.K.; Ramesh, V.; Palaniyandi, V.; Selvam, S.P.; Madhavan, T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J. Mol. Struct. 2020, 1221, 128823. [Google Scholar] [CrossRef] [PubMed]
- Yepes-Pérez, A.F.; Herrera-Calderon, O.; Quintero-Saumeth, J. Uncaria Tomentosa (cat’s Claw): A Promis. Herb. Med. SARS-CoV Junction SARS-CoV Spike Protein Based Mol. Model. J. Biomol. Struct. Dyn. 2020, 40, 2227–2243. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Sinha, S.K.; Prasad, S.K.; Islam, M.A.; Gurav, S.S.; Patil, R.B.; AlFaris, N.A.; Aldayel, T.S.; AlKehayez, N.M.; Wabaidur, S.M.; Shakya, A. Identification of bioactive compounds from Glycyrrhiza glabra Possible Inhib. SARS-CoV Spike Glycoprotein Non-Struct. Protein-15: A Pharmacoinformatics Study. J. Biomol. Struct. Dyn. 2020, 39, 4686–4700. [Google Scholar] [CrossRef]
- Hwang, S.S.; Lim, J.; Yu, Z.; Kong, P.; Sefik, E.; Xu, H.; Harman, C.C.D.; Kim, L.K.; Lee, G.R.; Li, H.B.; et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 2020, 367, 1255–1260. [Google Scholar] [CrossRef]
- Yi, Y.; Li, J.; Lai, X.; Zhang, M.; Kuang, Y.; Bao, Y.O.; Yu, R.; Hong, W.; Muturi, E.; Xue, H.; et al. Natural triterpenoids from licorice potently inhibit SARS-CoV-2 infection. J. Adv. Res. 2022, 36, 201–210. [Google Scholar] [CrossRef]
- Quimque, M.T.J.; Notarte, K.I.R.; Fernandez, R.A.T.; Mendoza, M.A.O.; Liman, R.A.D.; Lim, J.A.K.; Pilapil, L.A.E.; Ong, J.K.H.; Pastrana, A.M.; Khan, A.; et al. Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J. Biomol. Struct. Dyn. 2020, 39, 4316–4333. [Google Scholar] [CrossRef]
- Ghasemitarei, M.; Privat-Maldonado, A.; Yusupov, M.; Rahnama, S.; Bogaerts, A.; Ejtehadi, M.R. Effect of Cysteine Oxidation in SARS-CoV-2 Receptor-Binding Domain on Its Interaction with Two Cell Receptors: Insights from Atomistic Simulations. J. Chem. Inf. Model. 2021, 62, 129–141. [Google Scholar] [CrossRef]
- Allam, L.; Ghrifi, F.; Mohammed, H.; Hafidi, N.E.; Jaoudi, R.E.; Harti, J.E.; Lmimouni, B.; Belyamani, L.; Ibrahimi, A. Targeting the GRP78-Dependant SARS-CoV-2 Cell Entry by Peptides and Small Molecules. Bioinform. Biol. Insights 2020, 14, 117793222096550. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2020, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Yang, J.; Nune, M.; Zong, Y.; Zhou, L.; Liu, Q. Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP. Structure 2015, 23, 2191–2203. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE SC 2006 Conference (SC’06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar] [CrossRef]
- Amarelle, L.; Lecuona, E. The Antiviral Effects of Na,K-ATPase Inhibition: A Minireview. Int. J. Mol. Sci. 2018, 19, 2154. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; Chang, S.Y.; Byun, S.Y.; Ianevski, A.; Choi, I.; Pham Hung d’Alexandry d’Orengiani, A.L.; Ravlo, E.; Wang, W.; Bjørås, M.; Kainov, D.E.; et al. Screening of FDA-Approved Drugs Using a MERS-CoV Clinical Isolate from South Korea Identifies Potential Therapeutic Options for COVID-19. Viruses 2021, 13, 651. [Google Scholar] [CrossRef] [PubMed]
- Caohuy, H.; Eidelman, O.; Chen, T.; Liu, S.; Yang, Q.; Bera, A.; Walton, N.I.; Wang, T.T.; Pollard, H.B. Common cardiac medications potently inhibit ACE2 binding to the SARS-CoV-2 Spike, and block virus penetration and infectivity in human lung cells. Sci. Rep. 2021, 11, 22195. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, P.L.; Diener-West, M.; Callahan, K.A.; Lee, S.; Talbot, C.C.; Pollard, B.; Boyle, M.P.; Lechtzin, N. Digitoxin for Airway Inflammation in Cystic Fibrosis: Preliminary Assessment of Safety, Pharmacokinetics, and Dose Finding. Ann. Am. Thorac. Soc. 2017, 14, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, X.E. Construction and applications of SARS-CoV-2 pseudoviruses: A mini review. Int. J. Biol. Sci. 2021, 17, 1574–1580. [Google Scholar] [CrossRef] [PubMed]
- Gawish, R.; Starkl, P.; Pimenov, L.; Hladik, A.; Lakovits, K.; Oberndorfer, F.; Cronin, S.J.; Ohradanova-Repic, A.; Wirnsberger, G.; Agerer, B.; et al. ACE2 is the critical in vivo receptor for SARS-CoV-2 in a novel COVID-19 mouse model with TNF- and IFNγ-driven immunopathology. eLife 2022, 11, e74623. [Google Scholar] [CrossRef]
- Jamshidinia, N.; Mohammadipanah, F. Nanomaterial-Augmented Formulation of Disinfectants and Antiseptics in Controlling SARS CoV-2. Food Environ. Virol. 2022, 14, 105–119. [Google Scholar] [CrossRef]
- Luo, M.X.; Hua, S.; Shang, Q.Y. Application of nanotechnology in drug delivery systems for respiratory diseases. Mol. Med. Rep. 2021, 23, 325. [Google Scholar] [CrossRef]
- Rai, M.; Bonde, S.; Yadav, A.; Plekhanova, Y.; Reshetilov, A.; Gupta, I.; Golińska, P.; Pandit, R.; Ingle, A.P. Nanotechnology-based promising strategies for the management of COVID-19: Current development and constraints. Expert Rev. Anti-Infect. Ther. 2020, 1–10. [Google Scholar] [CrossRef]

















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iraci, N.; Corsaro, C.; Giofrè, S.V.; Neri, G.; Mezzasalma, A.M.; Vacalebre, M.; Speciale, A.; Saija, A.; Cimino, F.; Fazio, E. Nanoscale Technologies in the Fight against COVID-19: From Innovative Nanomaterials to Computer-Aided Discovery of Potential Antiviral Plant-Derived Drugs. Biomolecules 2022, 12, 1060. https://doi.org/10.3390/biom12081060
Iraci N, Corsaro C, Giofrè SV, Neri G, Mezzasalma AM, Vacalebre M, Speciale A, Saija A, Cimino F, Fazio E. Nanoscale Technologies in the Fight against COVID-19: From Innovative Nanomaterials to Computer-Aided Discovery of Potential Antiviral Plant-Derived Drugs. Biomolecules. 2022; 12(8):1060. https://doi.org/10.3390/biom12081060
Chicago/Turabian StyleIraci, Nunzio, Carmelo Corsaro, Salvatore V. Giofrè, Giulia Neri, Angela Maria Mezzasalma, Martina Vacalebre, Antonio Speciale, Antonina Saija, Francesco Cimino, and Enza Fazio. 2022. "Nanoscale Technologies in the Fight against COVID-19: From Innovative Nanomaterials to Computer-Aided Discovery of Potential Antiviral Plant-Derived Drugs" Biomolecules 12, no. 8: 1060. https://doi.org/10.3390/biom12081060