- 1State Key Laboratory of Desert and Oasis Ecology, Key Laboratory of Ecological Safety and Sustainable Development in Arid Lands, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Ürümqi, China
- 2Xinjiang Key Laboratory of Conservation and Utilization of Plant Gene Resources, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Ürümqi, China
- 3Turpan Eremophytes Botanical Garden, Chinese Academy of Sciences, Turpan, China
- 4State Key Laboratory of Crop Genetics and Germplasm Enhancement, MOA Key Laboratory of Plant Nutrition and Fertilization in Low-Middle Reaches of the Yangtze River, Nanjing Agricultural University, Nanjing, China
High-affinity K+ transporters (HKTs) are known as transmembrane cation transporters and are involved in Na+ or Na+-K+ transport in plants. In this study, a novel HKT gene SeHKT1;2 was isolated and characterized from the halophyte, Salicornia europaea. It belongs to subfamily I of HKT and shows high homology with other halophyte HKT proteins. Functional characterization of SeHKT1;2 indicated that it contributes to facilitating Na+ uptake in Na+-sensitive yeast strains G19, however, cannot rescue the K+ uptake-defective phenotype of yeast strain CY162, demonstrating SeHKT1;2 selectively transports Na+ rather than K+. The addition of K+ along with NaCl relieved the Na+ sensitivity. Furthermore, heterologous expression of SeHKT1;2 in sos1 mutant of Arabidopsis thaliana increased salt sensitivity and could not rescued the transgenic plants. This study will provide valuable gene resources for improving the salt tolerance in other crops by genetic engineering.
Introduction
Globally, soil salinity remains a significant abiotic stress for plants and constraints agriculture development. In order to solve the current problems of salt stress, we must understand how salt stress tolerance works and develop salt-tolerant crops. In addition to osmotic stress, high salinity can cause plant death and growth inhibition. A primary consequence of ionic stress is sodium disequilibrium in many plants, especially gramineous crops, which can adversely affect plant nutrition, enzyme activity, photosynthesis, and metabolism (Tester and Davenport, 2003; Mahajan and Tuteja, 2005). Na+ is attracted to the plasma membrane in saline environments due to high external Na+. A number of mechanisms are involved in plants acquiring salt tolerance, including the outright exclusion of Na+ and vacuolar compartmentalization. Na+ is extruded out of salt-stressed cells by plasma membrane Na+/H+ antiporters, also called SOS1 or NHA-type transporters. NHX-type antiporters, also known as Na+/H+ antiporters, sequestrate Na+ from the cytosol into vacuoles when salt stress is applied to the cell (Hasegawa et al., 2003; Zhu, 2003; Mahajan et al., 2008). In addition to controlling Na+ influx, salt tolerance can also be improved by reducing the amount of sodium absorbed. A number of studies have indicated that toxic Na+ influx into roots is mediated by nonselective cation channels (NSC) or voltage-independent channels (VIC) under high external Na+ concentrations (Amtmann and Sanders, 1998; Schachtman and Liu, 1999; Blumwald et al., 2000). In spite of this, NSC/VIC remains mysterious in terms of their detailed molecular identities. As a result, plant cells may be able to uptake Na+ through high-affinity K+ transporters (HKT-type transporters).
Based on amino acid in the first pore domain, the HKT transporters can be classified into two subclasses. Class I HKT transporters possess a serine (S) whereas members of class II have a glycine (G) residue at first pore forming region (Mäser et al., 2002b; Almeida et al., 2013). A single amino acid at this position determines the ion selectivity of the transporter. The class I HKT transporters showed preference for Na+ conductance over that of other cations whereas the class II HKT transporters select either Na+ and/or K+ (Mäser et al., 2002b; Almeida et al., 2014). A variety of plant species have been shown to have HKT-type transporters (Schachtman and Schroeder, 1994; Nieves-Cordones et al., 2016), and their functions have been studied in different ways. A heterologous expression system of TaHKT2;1 in wheat, for example, showed dual modes of action. In other words, when the Na+-K+ exchange rate was low, it acted as a Na+- K+ symporter, while when the exchange rate was high, it acted as a Na+-uniporter(Rubio et al., 1995; Gassmann et al., 1996). Transgenic wheat plants increased in growth under salinity when TaHKT2;1 was knocked down genetically (Laurie et al., 2002), indicating that TaHKT2;1 can be a Na+-uptake pathway in wheat roots. Several heterologous expression systems have also characterized Arabidopsis AtHKT1;1 and Rice OsHKT2;1 as Na+-uniporters (Uozumi et al., 2000; Horie et al., 2001; Garciadeblás et al., 2003). The genetic mutations of AtHKT1;1 in Arabidopsis roots showed that it does not mediate Na+ influx (Mäser et al., 2002a; Berthomieu et al., 2003; Horie et al., 2006); however, OsHKT2;1 mediates Na+ uptake in roots with low K+ levels (Horie et al., 2007). The salt-tolerance mechanisms of monocotyledonous halophytes can be used as a tool to improve salt tolerance crops.
It has been increasingly clear how halophytes have adapted to high salinity conditions over the past few decades. The morphological and biochemical adaptations of halophytes allow them to cope with high soil salinity (Flowers et al., 2010). Salicornia europaea is a salt marsh halophyte belonging to the Amaranthaceae family that is one of the most salt-tolerant plants on the planet(Patel, 2016). S. europaea imparted a higher salt tolerance. In S. europaea, salt tolerance may be attributed to its ability to restrict Na+ influx unidirectionally to roots, leading to a large concentration gradient between roots and shoots (Bressan et al., 2013). As the uptake of K+ is maintained in S. europaea roots, it has been proposed that the endodermal barrier to Na+ restricts Na+ uptake. The molecular identity of the channel or transporter associated with the restriction of unidirectional Na+ influx has not been disclosed (Xu et al., 2016).
In this study, we cloned a novel gene encoding a potassium transporter of the HKT type from S. europaea (SeHKT1;2). The gene expression pattern of SeHKT1;2 under K+-starvation and salt stress was analyzed and its function was characterized in yeast and Arabidopsis.
Materials and methods
Plant materials, growth conditions, and stress treatments
Seeds of S. europaea were collected from an alkaline soil area of Xinjiang, China. Plants were germinated in tap water for 14 days and then transferred to half-strength Hoagland’s nutrient solution and allowed to grow for another 14 days before stress treatments. The temperatures of the growth chamber were maintained at 28°C during the day (16 h) and 22°C at night (8 h) under a photoperiod (350-400 µmol m-2 s-1). For the salt-stress treatment, NaCl was added to half-strength Hoagland’s nutrient solution with 0 mM, 10 mM, 200 mM, 500 mM, and 800 mM separately. For the K+-starvation treatment, KCl was removed from the standard solution. Under 200 mM NaCl treatment, plants tissue samples were harvested at 0, 6, 24, 48, 72, and 120 h after stress treatments and preserved at –80°C for further analyses.
Cloning of SeHKT1;2 from S. europaea
Total RNA was extracted from the shoots of S. europaea using RNeasy plant extraction Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Total RNA was reverse transcribed by using an oligo dT primer and MMLV-reverse transcriptase (TaKaRa, China). Full-length cDNA was obtained using 5’- and 3’-RACE (Rapid Amplification of cDNA Ends) techniques. The partial cDNA fragment was amplified by PCR, using a degenerate primer (Table S1) deduced from the conserved regions of HKT and a reverse transcription product as a template. The full-length cDNA of SeHKT1;2 was amplified using primers (Table S1). The PCR product was sub-cloned into the pMD-19 T vector and sequenced.
Bioinformatic analysis of SeHKT1;2 sequence
Transmembrane domains were predicted by DeepTMHMM (https://biolib.com/damgaard/DeepTMHMM-testing). A maximum-likelihood phylogenetic tree of plant HKT proteins was constructed using the MEGA program (version 7.0, Auckland, NewZealand). The accession numbers of genes utilized in the present study are listed in Table S2. The protein secondary structure was predicted using NetSurfP - 3.0 online program(Høie et al., 2022). The 3D model of SeHKT1;2 protein was build using SWISS-MODEL online program (Waterhouse et al., 2018).
RNA extraction and cDNA synthesis for real-time RT-PCR
For real-time RT-PCR, total RNA was extracted from the shoots and roots of S. europaea subjected to 0 mM Na+, 10 mM Na+, 200 mM Na+, 500 mM Na+, and 800 mM Na+, K+-starvation, time-course using the RNeasy plant extraction Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Two micrograms of DNase-treated RNA were transcribed to cDNA using the Reversal Transcription Reagent Kit (TaKaRa) following the manufacturer’s instructions. The cDNA was diluted 10 times and 1µl of the diluted cDNA was used as the template in each well for quantitative real-time PCR analysis. The cDNA was amplified using Power SYBR Green PCR Master Mix (Applied Biosystems, USA) on CFX96 Real-Time PCR Detection System (Bio-Rad, USA). A α-tubulin gene from S. europaea has been used as internal reference (Lv et al., 2012; Ma et al., 2013; Xiao et al., 2015) and served as an internal standard to normalize the expression data for the SeHKT1;2 gene. The qRT-PCR was performed using sequence specific primers (Table S2). The thermal profile for qRT-PCR was as follows: 2 min at 95°C, 40 cycles of 15 s at 95°C and 30 s at 60°C, and a melting curve protocol (plates read when increased 0.5°C every 5 s from 65°C to 95°C). The melting curve verified the amplification specific and confirmed that there were no primer dimers. All of the samples were run with replicates. The threshold cycle (Ct) values were measured according to the setting of an auto-calculated baseline threshold in Bio-Rad CFX Manager software (Bio-Rad, USA).
Functional characterization of SeHKT1;2 in yeast
The ORF of SeHKT1;2 was inserted into the yeast protein expression vector pYES2. To test the Na+ Absorption and efflux function of SeHKT1;2, the Saccharomyces cerevisiae yeast mutant strain AXT3K (enal::Hls3::ena4,nhal::LEuZ,nhxl::KanMx4), which lacks the main plasma membrance Na+ transporters, and G19 (MATa ade2ura3leu2his3trp1 ena1Δ::HIS3Δ::ena4Δ) disrupted in the ENA1-4 genes encoding Na+ export pumps were used. The plasmids were introduced by PEG/LiAc method. Positive transformants were selected on Ura-selective medium [0.67% (w/v) yeast nitrogen base without amino acids, 0.077% (w/v) DO supplement-Ura, 2% (w/v) galactose, and 1.5% (w/v) agar]. Growth at variable Na+ concentrations with 0, 25, 30, 60, 100 or 150 mM for AXT3K and 0, 60, 100, 150, 300 mM for G19 were tested in arginine phosphate (AP) medium [8 mM phosphoric acid, 10 mM L-Arginine, 2 mM MgSO4, 0.2 mM CaCl2, 2% glucose, plus vitamins and trace elements, and 1.5% (w/v) agar, pH 6.5].
Meanwhile, Yeast (Saccharomyces cerevisiae) strain CY162 (MATa, △trk1, trk2:: pCK64, his3, leu2, ura3, trp1, ade2), which is a K+-uptake-defective mutant and cannot grow without supplement K+ was used to test K+-uptake function of SeHKT1;2 gene. Positive transformants were selected on Ura-selective medium with 100 mM KCl. Yeast growth experiments were performed on arginine-phosphate (AP) medium with added K+ (1 mM, 10 mM) and supplemented with 100 mM, 150 mM and 300 mM Na+ concentrations. Control experiments were performed with the yeast modified with vector pYES2 and pYES2-SeHKT1;2 growing under 100 mM KCl.
For the yeast growth test experiment, all transformed yeasts were cultured overnight at 30°C in AP medium until the OD600 reached 0.8, and 10-fold serial diluted cultures were incubated on AP plates containing the indicated concentrations of K+ and Na+. The plates were incubated at 30°C for 5 days. Control experiments were performed with the yeast wild type modified with vector pYES2.
Generation of transgenic Arabidopsis plants over-expressing the SeHKT1;2 gene
The coding regions of SeHKT1;2 was sub-cloned into the plant transformation binary vector pBI121 (Clontech, Japan) under the control of CaMV35S promoters. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 and Arabidopsis salt-hypersensitive mutant sos1 was transformed by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength Murashige & Skoog medium containing 25 mg/L kanamycin. qRT-PCR showed the introduced gene transcript in 10 out of 15 lines of SeHKT1;2 with varied expression levels among the lines. Two lines transgenic plants with higher expression level were used for phenotype assays of salt stresses. All of the lines used in these experiments were homozygous. Wild type and transgenic Arabidopsis seeds were surface-sterilized with 70% (v/v) ethanol for 2 min and 1% (v/v) NaCl solution for 15 min. The seeds were rinsed three times with water and sown on an MS medium containing 1.5% (w/v) sucrose, 0.8% (w/v) agar, and 50 mg/L kanamycin. Seven-day-old seedlings were transferred from germination medium to soil, supplemented with the indicated amounts of 300 mM NaCl after ten days.
Results
Identification of SeHKT1-like gene from S. europaea
By using degenerate primers deduced from several HKT sequences and standard reverse transcription (RT)-PCR methods, a cDNA homolog of an HKT high-affinity K+ transporter from S. europaea was cloned. The length of the amplified cDNA fragment was about 1500-bp long. The full-length cDNA was obtained by 5′ and 3′ RACE (Rapid Amplification of cDNA Ends) which contains an ORF of 1611-bp long (Figure S1A). The translated amino acid sequence of this gene exhibited 44.31% amino acid sequence identity with SeHKT1;1 (AKS12114.1) (Figure S1B) therefore it was designated as SeHKT1;2 (AKS2645929). Subsequently the evolutionary relationships of SeHKT1;2 with HKT proteins from other plant species. An unrooted phylogenetic tree was generated based on full-length amino acid sequences. Results showed that SeHKT1;2 was grouped into subfamily I of HKT transporters (Figure 1).

Figure 1 The phylogenetic relationship of SeHKT1;2 with HKTs from other plant species. Full length of HKT proteins form different plant species were used and the tree was generated by using neighbor-joining method with 1000 bootstrap replicates. Accession numbers and species for all sequences are listed in Supplemental Table S1.
Structural characteristic of SeHKT1;2
The SeHKT1;2 encoding a 536 amino acid polypeptide with 8 predicted transmembrane domains (Figure 2). SMART domain architecture analysis showed that SeHKT1;2 protein has single TrkH conservative domain (pfam ID: PF02386) between amino acids 70-524 (Figure S2A). The secondary structure of the SeHKT1;2 protein was predicted, and it was found to have a complex helical folding structure (Figure S2B). The 3D models of SeHKT1;2 showed the presence of three glycine residues (Gly244, Gly368 and Gly480) and one serine residue (Ser119) forming an ion selectivity filter, which belong to subfamily I (Figures 2, S3).
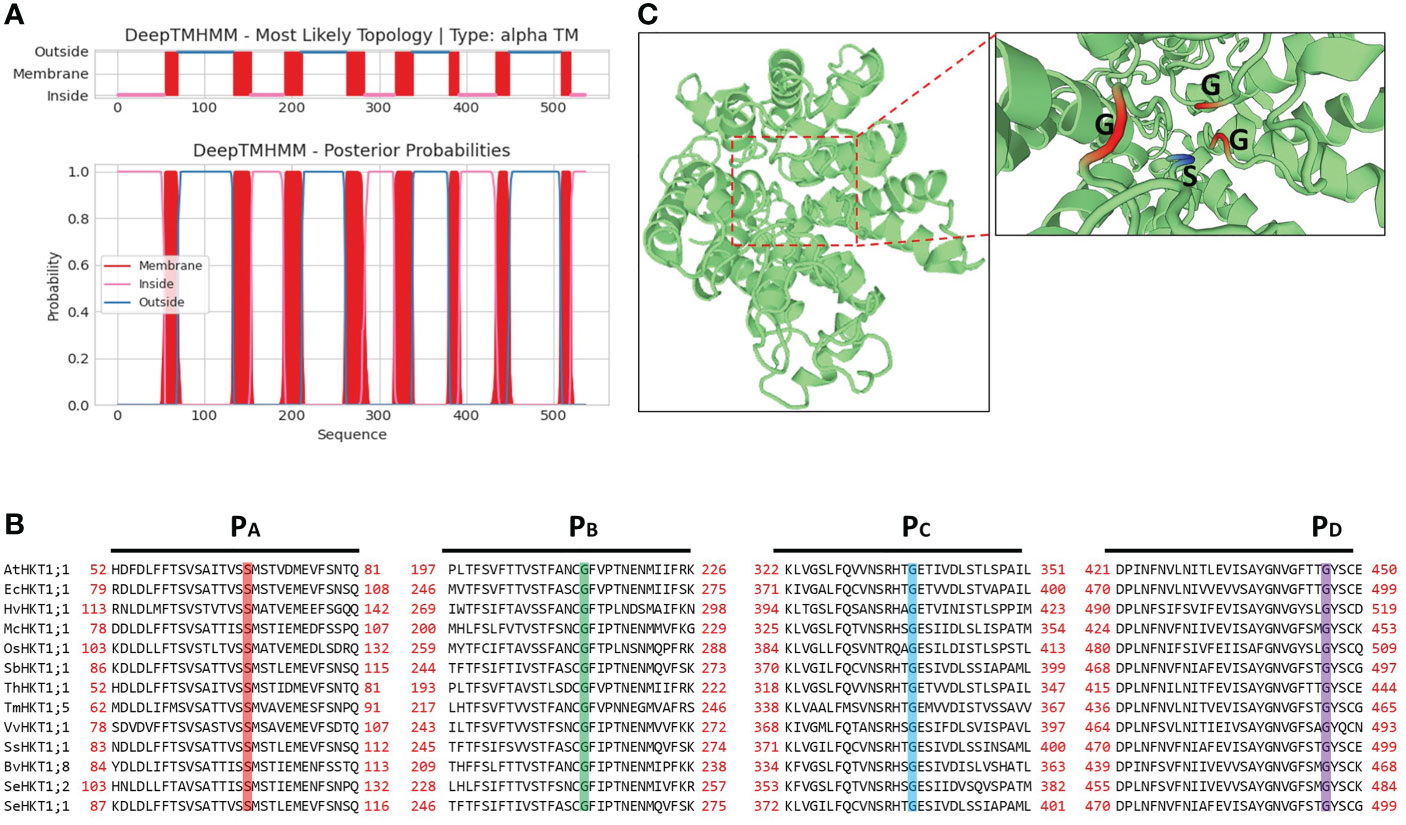
Figure 2 Structure analysis of SeHKT1;2 protein. (A) Prediction of the transmembrane domain of SeHKT1;2 protein. Blue lines are pore loop regions, and red boxes are transmembrane regions. (B) Multiple sequence alignment of the four conserved selectivity-filter-pore regions of HKT proteins. Amino acid sequences were aligned using Clustal W and visualized using BioEdit. Conserved residues in pore loops were indicated with colored lines. (C) Predicted 3D model of SeHKT1;2 protein. The first P-loop (PA, S/G) and the other P-loop (PB-D, G) are shown in magenta and orange (Color figure online).
Tissue specific expression of SeHKT1;2 gene
The expression pattern of SeHKT1;2 gene under the salt stress conditions was determined. The SeHKT1;2 gene expression patterns in S. europaea seedlings were completely different between shoots and roots. In a time-course experiment, the SeHKT1;2 was induced by NaCI (200mM) and reached the peak at 48h in shoot, but it was gradually reduced in root (Figure 3). Subsequently, we quantified expression level of SeHKT1;2 gene under the presence of NaCI. As a result, the 10 mM NaCI significantly increased the expression of SeHKT1;2 in shoot, but it remained same in the presence of high concentration (200mM, 500mM, 800 mM) of NaCI (Figure 3). In addition, we also examined the SeHKT1;2 gene expression under K+ starved condition. In our experiments, SeHKT1;2 showed a very low expression level in roots while showed 10 folds higher in shoots (Figure 3). The expression of SeHKT1;2 in root was not affected by NaCI.
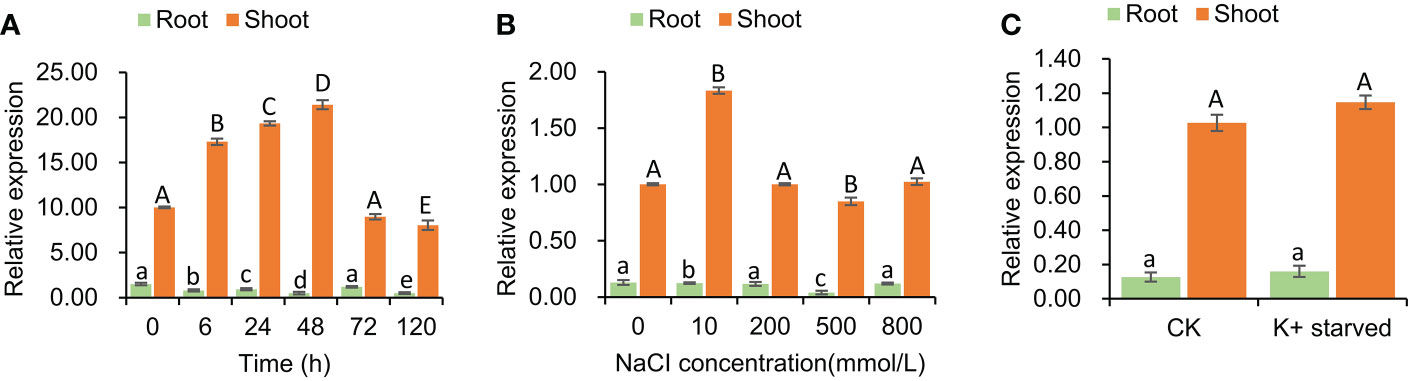
Figure 3 Tissue specific expression of SeHKT1;2 gene. (A) Time course expression level of SeHKT1;2 gene in shoots and roots. (B) The relative expression level of SeHKT1;2 gene under 0, 10, 200, 500, 800 mmol NaCl concentration treatment. Plant samples were taken after 72h of NaCl treatment. (C) Expression pattern of SeHKT1;2 gene under K+ starved condition. After the seedlings were pre-cultured with standard Hoagland nutrient solution for 30 days, the seedlings were divided two groups. One group were continue cultured with standard Hoagland nutrient solution as CK group. Another group were cultured with - free Hoagland nutrient as K+-starved group. Plant samples were taken after 72h of K+ starvation treatment. α-tubulin was used as the internal reference for data processing. Values indicated the mean and standard deviation of biological repetitions (n = 3).
Functional characterization of SeHKT1;2 in yeast
Previous functional characterization in yeasts showed that SaHKT1 from the Suaeda salsa functions as a Na+ selective transporter (Wang et al., 2020). To determine whether SeHKT1;2 is involved in Na+ uptake, we expressed SeHKT1;2 proteins in a Na+-sensitive mutant yeast strain, G19 (Quintero et al., 1996). Results showed that under the low concentration of K+ (1Mm), the increasing of Na+ concentration in the growth medium caused severe growth defects in both SeHKT1;2-expressing cells and empty vector-harboring control cells (Figure 4 left panel). Interestingly, higher K+ concentration (10mM) improved the growth of both control and SeHKT1;2-expressing cells (Figure 4 right panel). Moreover, the higher concentration (300mM) of Na+ restricted cell growth of both control and SeHKT1;2-expressing cells (Figure S4). These data indicate that Na+ toxicity was reduced by K+ uptake, and that cells expressing SeHKT1;2 has a selective advantage under K+ limitation suggesting that SeHKT1;2 have a higher affinity for K+.
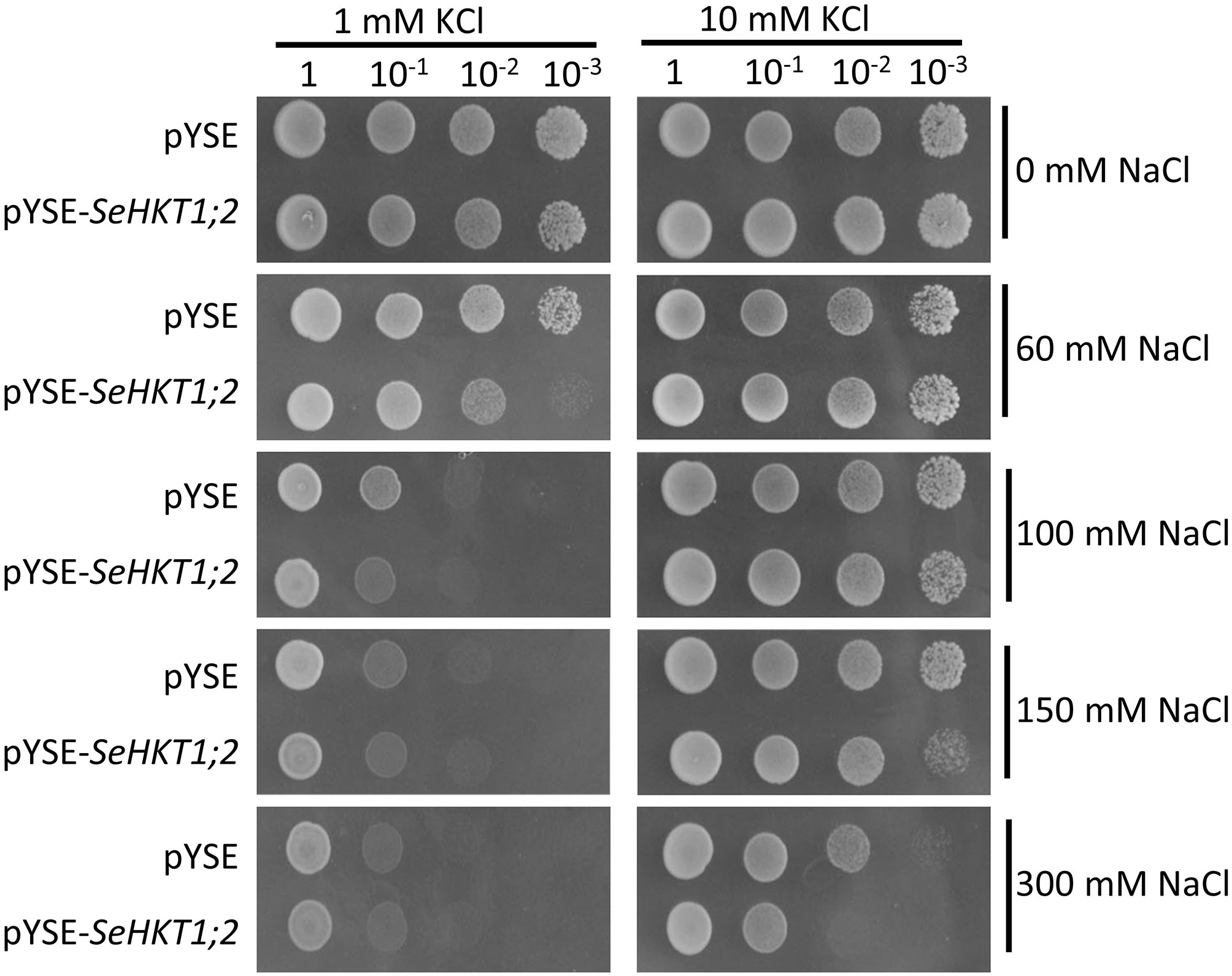
Figure 4 Functional characterizations of SeHKT1;2 in G19 yeast strain. Growth of G19 yeast cells transformed by empty vector (pYSE) control and pYSE-SeHKT1;2. Each G19 transformants on solid arginine phosphate (AP) medium supplemented with various concentrations (1mM,10 mM) of KCl with combination of different salt concentrations (0, 60, 100, 150, and 300 mM NaCl) were incubated at 30°C for 3 days. Numbers (1, 10-1, 10-2, 10-3) on the top of each panel indicated serial dilutions of yeast cells placed on the medium.
To investigate the transporter activity of SeHKT1;2, we performed complementation assay by ectopic expression of SeHKT1;2 in the K+ transporter-deficient yeast strain CY162 (Anderson et al., 1992). We observed no significant differences between SeHKT1;2 and empty vector control in their growth supplied with various concentration of KCl, and both grew well on the medium containing 100 mM KCI (Figure 5A). This indicates that SeHKT1;2 failed to complement K+ uptake. Additionally, the transformants growth was completely impacted as the non-transformed strain when K+ and Na+ were applied in combined form, in the CY162 with increased K+ (10Mm KCI) in combination with Na+, the transformants growth was somehow recovered (Figure 5B). These results suggested that a trend toward K+ selectivity by SeHKT1;2.
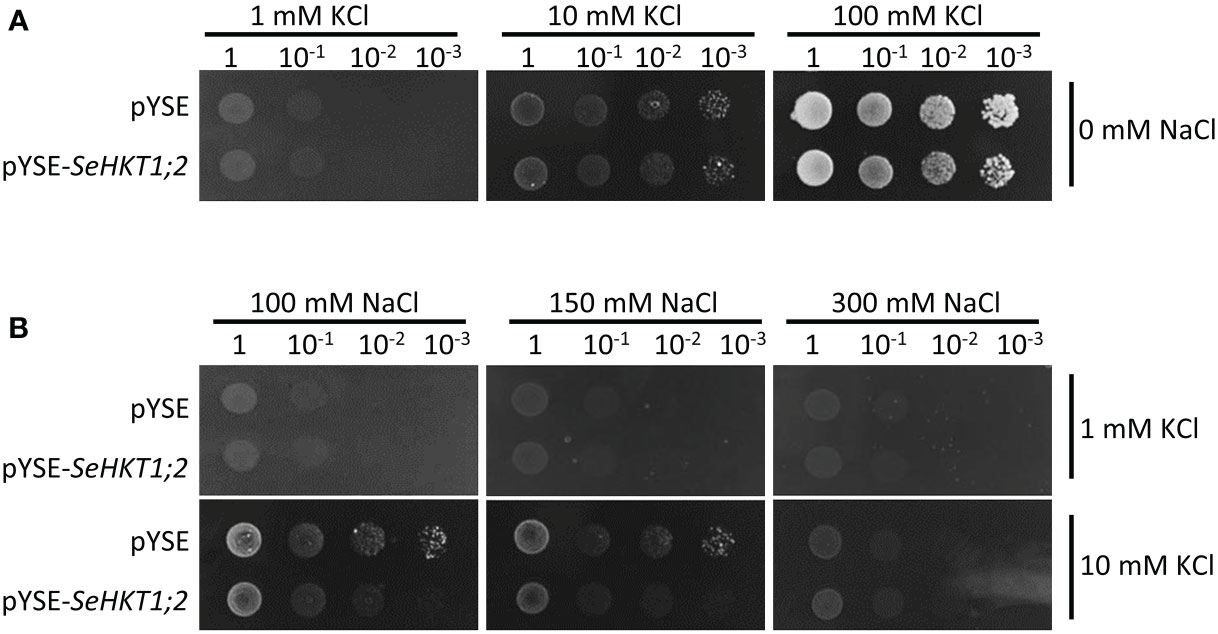
Figure 5 Complementation assay in yeast CY162 strain. (A) Growth of yeast strain CY162 cells harboring the empty vector (pYSE) control and pYSE-SeHKT1;2. Growth of each CY162 transformant on solid arginine phosphate (AP) medium supplemented with various concentrations (1mM,10 mM, 100 mM) of KCl and 0 mM NaCl incubated at 30°C for 3 days. Numbers (1, 10-1, 10-2, 10-3) on the top of each panel indicated serial dilutions of yeast cells placed on the medium. (B) Growth status of the yest cell lines in (A) on AP medium containing different concentration of KCl (1 mM,10 mM) and NaCl (100mM,150mM,300mM). Each yest cell lines were incubated at 30°C for 3 days. Numbers (1, 10-1, 10-2, 10-3) on the top of each panel indicated serial dilutions of yeast cells placed on the medium.
Heterologous expression of SeHKT1;2 in Arabidopsis sos1 mutant
In an attempt to validate the function of SeHKT1;2 in salt tolerance, we over expressed the SeHKT1;2 in Arabidopsis sos1 mutants. As expected, the whole phenotypes of SeHKT1;2-expressing in sos1 and wild type Col-0 plants showed the same growth vigor and had no obvious difference under the normal condition. The growth of sos1 plant, however, was restricted by imposing of salt stresses (300 mM NaCl) and attenuated by overexpression of SeHKT1;2 (Figure 6). Interestingly, the overexpression of SeHKT1;2 cannot restore the salt tolerance of sos1 mutant plant.

Figure 6 Phenotypes of transgenic and wild type Arabidopsis lines under salt stress. The sos1 mutant Arabidopsis were infiltrated by Agrobacterium tumefaciens harbored the SeHKT1;2 construct. Four-week-old Arabidopsis plants were treated with watered with 300 mM NaCl for 14 days.
Discussion
HKT transporters are one of the well-studied cation transporters in plant species. From an S. europaea halophyte, a homolog of HKT, SeHKT1;2, was isolated and the amino acids at the four pore-loop conservative sites of SeHKT1;2 protein are Ser-Gly-Gly-Gly, which belong to subfamily I (Figure 2). The presence of a Ser (serine) in the first pore-lope regions of HKT protein is associated with Na+ selectivity, and is associated with Na+ and/or K+ selectivity only when the serine at this position is changed to Gly (glycine) (Horie et al., 2009). Although the rice OsHKT2;1 has Ser residue at the filter position of the first P-loop but exhibits features of class I transporters such as poor K+ permeability (Horie et al., 2001; Yao et al., 2010).
The AtHKT1, EsHKT1;2(TsHKT1;2) and EpHKT1;2 belong to subfamily I and contain the conserved serine residue in the first pore-lope region (Ali et al., 2012; Ali et al., 2018). Unlike the AtHKT1, the EsHKT1;2 and EpHKT1;2 show significantly higher affinity for K+ than for Na+ (Ali et al., 2018). Presence of aspartic acid residue (D) in the second pore-loop domain of EpHKT1;2 (D207,D238) and EsHKT1;2 (D205,D236) determines their cation specificity (Na+ or K+) (Ali et al., 2016; Ali et al., 2018) suggesting that glycine residues are not the only amino acids involved in K+ transport in HKT proteins. SeHKT1;2 contains asparagine residue (N) instead of aspartic acid residue (D) in the second pore-loop domain at the corresponding positions (D242, D273) in which same with AtHKT1 (Figure S5) and supposed to has functional similarity with AtHKT1. In contrast, SeHKT1;2 exhibited higher affinity for K+ (Figures 4, 5), whereas AtHKT1 showed a higher affinity for Na+. Although SeHKT1;2 shared similarity with AtHK1 in key amino acids but functionally different form AtHK1 indicating that other structural differences, presently unknown, may enhance K+ uptake of SeHKT1;2.
Plant HKT gene expression is always affected by salt stresses. PtHKT1;5 from a halophytic grass, Puccinellia tenuiflora, was up regulated in root cells by salt (NaCI) stress. Unlike PtHKT1;5, the SsHKT1;1 from another halophyte, Suaeda salsa, was downregulated under NaCl treatment. It seems that the expression of HKT1 is up-regulated in salt-excluding while down-regulated salt-accumulating halophyte species under salt stress. S. europaea is a salt-accumulating halophyte, and transcript of SeHKT1;2 in root was also decreased by the time under salt stress (Figure 3). In contrast, salt stress induced the SeHKT1;2 gene expression in shoot of S. europaea (Figure 3). The differential expression of SeHKT1;2 gene in root and shoot may reduce Na+ retrieval from the xylem and facilitate Na+ transport into its shoots so S. europaea could adjust to high external salinity and exhibit better growth under saline environment. Hence, SeHKT1;2 is key protein in salt accumulation in S. europaea.
Plants maintain sodium homeostasis by Salt Overly Sensitive (SOS) pathway (Yang and Guo, 2018). AtSOS1 is the key sodium transporter in Arabidopsis SOS pathway. The sos1 mutant Arabidopsis plants are even more sensitive to Na+ stresses compared with sos3 mutant plants (Zhu et al., 1998; Shi et al., 2000). A previous study demonstrated that Na+ efflux rate significantly decreased by 16% in sos1 compared with WT (Wang et al., 2019). In our study, sos1 exhibited more sensitivity to salt stress and heterologous expression of SeHKT1;2 could not rescue the sos1 mutant plants but increased the sensitivity of the sos1 mutant plants (Figure 6). In the sos1 mutant plants the impairment of AtSOS1 mediated efflux of cytoplasmic Na+ resulting in accumulation of cytoplasmic Na+ and showed salt sensitivity (Figures 6, 7). The salt overly sensitive phenotype caused by heterologous expression of SeHKT1;2 can be explained that the SeHKT1;2 in sos1 Arabidopsis influx Na+ so that further accumulate cytoplasmic Na+ and eventually inhibits plant growth and increases salt sensitivity of sos1 plant (Figures 6, 7). This study suggested that SeHKT1;2 play very important roles in synergistically regulating Na+ homeostasis by controlling Na+ transport in Arabidopsis.
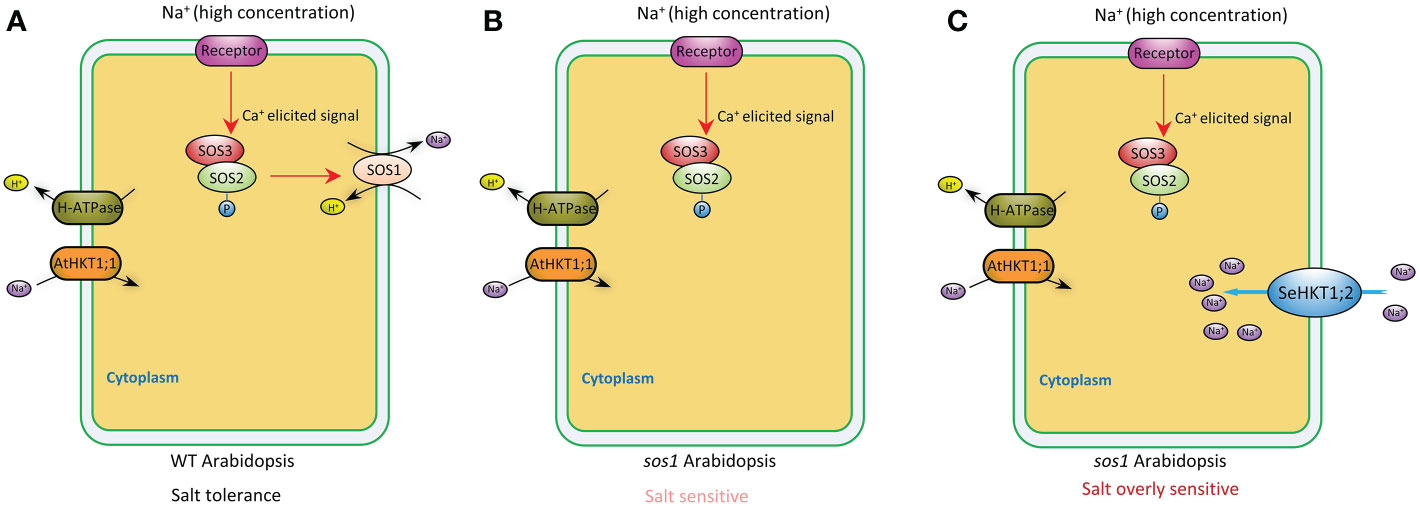
Figure 7 A proposed model depicting the putative function of SeHKT1;2 contributing to Na+ uptake across the plasma membrane. In wild type Arabidopsis, high Na+ concentration elicits a Ca2+ signal that activates SOS3-SOS2 complex, which in turn stimulates cytoplasmic Na+ efflux activity of SOS1 across the plasma membrane (A). Efflux of cytoplasmic Na+ was reduced in sos1 Arabidopsis, making the plants salt sensitive (B). Na+ uptake across the plasma membrane by SeHKT1;2 increased the cytoplasmic Na+ concentration, making the plants salt overly sensitive (C).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
The experimental design, data analyzation, manuscript organization were completed by JM and YH and ZP were assistant with RNA quantification. JM and LW conceived the project, supervised the analysis, and critically revised the manuscript. XF revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Xinjiang Uygur Autonomous Region regional coordinated innovation project (Shanghai cooperation organization science and technology partnership program) (No. 2022E01022), Introduction Project of High-level Talents in Xinjiang Uygur Autonomous Region.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1104070/full#supplementary-material
References
Ali, A., Khan, I. U., Jan, M., Khan, H. A., Hussain, S., Nisar, M., et al. (2018). The high-affinity potassium transporter EpHKT1;2 from the extremophile eutrema parvula mediates salt tolerance. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01108
Ali, Z., Park, H. C., Ali, A., Oh, D.-H., Aman, R., Kropornicka, A., et al. (2012). TsHKT1;2, a HKT1 homolog from the extremophile arabidopsis relative thellungiella salsuginea, shows k+ specificity in the presence of NaCl. Plant Physiol. 158, 1463–1474. doi: 10.1104/pp.111.193110
Ali, A., Raddatz, N., Aman, R., Kim, S., Park, H. C., Jan, M., et al. (2016). A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol. 171, 2112–2126. doi: 10.1104/pp.16.00569
Almeida, P. M. F., de Boer, G.-J., de Boer, A. H. (2014). Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SlHKT1;2 expressed in xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00600
Almeida, P., Katschnig, D., de Boer, A. H. (2013). HKT transporters–state of the art. Int. J. Mol. Sci. 14, 20359–20385. doi: 10.3390/ijms141020359
Amtmann, A., Sanders, D. (1998). Mechanisms of na+ uptake by plant cells. Adv. Bot. Res. 29, 75–112. doi: 10.1016/S0065-2296(08)60310-9
Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., Gaber, R. F. (1992). Functional expression of a probable arabidopsis thaliana potassium channel in saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 89, 3736. doi: 10.1073/PNAS.89.9.3736
Berthomieu, P., Conéjéro, G., Nublat, A., Brackenbury, W. J., Lambert, C., Savio, C., et al. (2003). Functional analysis of AtHKT1 in arabidopsis shows that na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 22, 2004–2014. doi: 10.1093/EMBOJ/CDG207
Blumwald, E., Aharon, G. S., Apse, M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465, 140–151. doi: 10.1016/S0005-2736(00)00135-8
Bressan, R. A., Park, H. C., Orsini, F., Oh, D. h., Dassanayake, M., Inan, G., et al. (2013). Biotechnology for mechanisms that counteract salt stress in extremophile species: A genome-based view. Plant Biotechnol. Rep. 7, 27–37. doi: 10.1007/S11816-012-0249-9/FIGURES/3
Clough, S. J., Bent, A. F. (1998). Floral dip: A simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/J.1365-313X.1998.00343.X
Flowers, T. J., Galal, H. K., Bromham, L. (2010). Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 37, 604–612. doi: 10.1071/FP09269
Garciadeblás, B., Senn, M. E., Bañuelos, M. A., Rodríguez-Navarro, A. (2003). Sodium transport and HKT transporters: The rice model. Plant J. 34, 788–801. doi: 10.1046/J.1365-313X.2003.01764.X
Gassmann, W., Rubio, F., Schroeder, J. I. (1996). Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 10, 869–882. doi: 10.1046/J.1365-313X.1996.10050869.X
Høie, M. H., Kiehl, E. N., Petersen, B., Nielsen, M., Winther, O., Nielsen, H., et al. (2022). NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 50, W510–W515. doi: 10.1093/NAR/GKAC439
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2003). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/ANNUREV.ARPLANT.51.1.463
Horie, T., Costa, A., Kim, T. H., Han, M. J., Horie, R., Leung, H. Y., et al. (2007). Rice OsHKT2;1 transporter mediates large na+ influx component into k+-starved roots for growth. EMBO J. 26, 3003–3014. doi: 10.1038/SJ.EMBOJ.7601732
Horie, T., Hauser, F., Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660. doi: 10.1016/J.TPLANTS.2009.08.009
Horie, T., Horie, R., Chan, W. Y., Leung, H. Y., Schroeder, J. I. (2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47, 622–633. doi: 10.1093/PCP/PCJ029
Horie, T., Yoshida, K., Nakayama, H., Yamada, K., Oiki, S., Shinmyo, A. (2001). Two types of HKT transporters with different properties of na+ and k+ transport in oryza sativa. Plant J. 27, 129–138. doi: 10.1046/J.1365-313X.2001.01077.X
Laurie, S., Feeney, K. A., Maathuis, F. J. M., Heard, P. J., Brown, S. J., Leigh, R. A. (2002). A role for HKT1 in sodium uptake by wheat roots. Plant J. 32, 139–149. doi: 10.1046/J.1365-313X.2002.01410.X
Lv, S., Jiang, P., Chen, X., Fan, P., Wang, X., Li, Y. (2012). Multiple compartmentalization of sodium conferred salt tolerance in salicornia europaea. Plant Physiol. Biochem. 51, 47–52. doi: 10.1016/J.PLAPHY.2011.10.015
Ma, J., Zhang, M., Xiao, X., You, J., Wang, J., Wang, T., et al. (2013). Global transcriptome profiling of salicornia europaea l. shoots under NaCl treatment. PloS One 8, e65877. doi: 10.1371/JOURNAL.PONE.0065877
Mahajan, S., Pandey, G. K., Tuteja, N. (2008). Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi: 10.1016/J.ABB.2008.01.010
Mahajan, S., Tuteja, N. (2005). Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/J.ABB.2005.10.018
Mäser, P., Eckelman, B., Vaidyanathan, R., Horie, T., Fairbairn, D. J., Kubo, M., et al. (2002a). Altered shoot/root na+ distribution and bifurcating salt sensitivity in arabidopsis by genetic disruption of the na+ transporter AtHKT1. FEBS Lett. 531, 157–161. doi: 10.1016/S0014-5793(02)03488-9
Mäser, P., Hosoo, Y., Goshima, S., Horie, T., Eckelman, B., Yamada, K., et al. (2002b). Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl. Acad. Sci. U. S. A. 99, 6428. doi: 10.1073/PNAS.082123799
Nieves-Cordones, M., Ródenas, R., Chavanieu, A., Rivero, R. M., Martinez, V., Gaillard, I., et al. (2016). Uneven HAK/KUP/KT protein diversity among angiosperms: Species distribution and perspectives. Front. Plant Sci. 7. doi: 10.3389/FPLS.2016.00127/BIBTEX
Patel, S. (2016). Salicornia: Evaluating the halophytic extremophile as a food and a pharmaceutical candidate. 3 Biotech. 6. doi: 10.1007/S13205-016-0418-6
Quintero, F. J., Garciadeblás, B., Rodríguez-Navarro, A. (1996). The SAL1 gene of arabidopsis, encoding an enzyme with 3’(2’),5’-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8, 529–537. doi: 10.1105/TPC.8.3.529
Rubio, F., Gassmann, W., Schroeder, J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. doi: 10.1126/SCIENCE.270.5242.1660
Schachtman, D., Liu, W. (1999). Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 4, 281–287. doi: 10.1016/S1360-1385(99)01428-4
Schachtman, D. P., Schroeder, J. I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658. doi: 10.1038/370655A0
Shi, H., Ishitani, M., Kim, C., Zhu, J.-K. (2000). The arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97, 6896–6901. doi: 10.1073/pnas.120170197
Tester, M., Davenport, R. (2003). Na+ tolerance and na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/AOB/MCG058
Uozumi, N., Kim, E. J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., et al. (2000). The arabidopsis HKT1 gene homolog mediates inward na(+) currents in xenopus laevis oocytes and na(+) uptake in saccharomyces cerevisiae. Plant Physiol. 122, 1249–1259. doi: 10.1104/PP.122.4.1249
Wang, Q., Guan, C., Wang, P., Ma, Q., Bao, A.-K., Zhang, J.-L., et al. (2019). The effect of AtHKT1;1 or AtSOS1 mutation on the expressions of na+ or k+ transporter genes and ion homeostasis in arabidopsis thaliana under salt stress. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20051085
Wang, W. Y., Liu, Y. Q., Duan, H. R., Yin, X. X., Cui, Y. N., Chai, W. W., et al. (2020). SsHKT1;1 is coordinated with SsSOS1 and SsNHX1 to regulate na+ homeostasis in suaeda salsa under saline conditions. Plant Soil 449, 117–131. doi: 10.1007/S11104-020-04463-X/FIGURES/10
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/NAR/GKY427
Xiao, X., Ma, J., Wang, J., Wu, X., Li, P., Yao, Y. (2015). Validation of suitable reference genes for gene expression analysis in the halophyte salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00788
Xu, C., Tang, X., Shao, H., Wang, H. (2016). Salinity tolerance mechanism of economic halophytes from physiological to molecular hierarchy for improving food quality. Curr. Genomics 17, 207. doi: 10.2174/1389202917666160202215548
Yang, Y., Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yao, X., Horie, T., Xue, S., Leung, H. Y., Katsuhara, M., Brodsky, D. E., et al. (2010). Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol. 152, 341. doi: 10.1104/PP.109.145722
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445. doi: 10.1016/S1369-5266(03)00085-2
Keywords: Salicornia europaea, salt tolerance, high-affinity K+ transporters, HKT, ion selectivity
Citation: Haxim Y, Wang L, Pan Z, Fan X and Ma J (2023) A novel high-affinity potassium transporter SeHKT1;2 from halophyte Salicornia europaea shows strong selectivity for Na+ rather than K+. Front. Plant Sci. 14:1104070. doi: 10.3389/fpls.2023.1104070
Received: 21 November 2022; Accepted: 06 February 2023;
Published: 20 February 2023.
Edited by:
Jin-Lin Zhang, Lanzhou University, ChinaReviewed by:
Haijun Gong, Northwest A&F University, ChinaMohamed Magdy F. Mansour, Ain Sham University, Egypt
Copyright © 2023 Haxim, Wang, Pan, Fan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbiao Ma, majinbiao@ms.xjb.ac.cn; Xiaorong Fan, xiaorongfan@njau.edu.cn
 Yakupjan Haxim
Yakupjan Haxim Lei Wang
Lei Wang Zhendong Pan1,3
Zhendong Pan1,3 Xiaorong Fan
Xiaorong Fan Jinbiao Ma
Jinbiao Ma