- 1Center for Host Defense, Inflammation, and Lung Disease (CHILD) Research, Department of Pediatrics, The Pennsylvania State University College of Medicine, Hershey, PA, United States
- 2Department of Obstetrics & Gynecology, The Pennsylvania State University College of Medicine, Hershey, PA, United States
- 3Department of Biochemistry & Molecular Biology, The Pennsylvania State University College of Medicine, Hershey, PA, United States
- 4School of Science, Engineering, and Technology, The Pennsylvania State University, Harrisburg, PA, United States
The human innate host defense molecules, SP-A1 and SP-A2 variants, differentially affect survival after infection in mice and in lung transplant patients. SP-A interacts with the sentinel innate immune cell in the alveolus, the alveolar macrophage (AM), and modulates its function and regulation. SP-A also plays a role in pulmonary surfactant-related aspects, including surfactant structure and reorganization. For most (if not all) pulmonary diseases there is a dysregulation of host defense and inflammatory processes and/or surfactant dysfunction or deficiency. Because SP-A plays a role in both of these general processes where one or both may become aberrant in pulmonary disease, SP-A stands to be an important molecule in health and disease. In humans (unlike in rodents) SP-A is encoded by two genes (SFTPA1 and SFTPA2) and each has been identified with extensive genetic and epigenetic complexity. In this review, we focus on functional, structural, and regulatory differences between the two SP-A gene-specific products, SP-A1 and SP-A2, and among their corresponding variants. We discuss the differential impact of these variants on the surfactant structure, the alveolar microenvironment, the regulation of epithelial type II miRNome, the regulation and function of the AM, the overall survival of the organism after infection, and others. Although there have been a number of reviews on SP-A, this is the first review that provides such a comprehensive account of the differences between human SP-A1 and SP-A2.
1. Introduction
The key function of the lung is the exchange of O2 and CO2, which is vital for the survival of the organism. This function occurs in the distal lung air spaces called alveoli. The alveolus is lined by epithelial cells, the type I cells, constituting the barrier for the CO2/O2 exchange and the type II cells, the cellular site of surfactant production. The alveoli are lined by a thin liquid layer, the hypophase (Figure 1A). Pulmonary surfactant, a lipoprotein complex, is found at the air-liquid interface of the alveolus, as well as in the hypophase, and is essential for life. By lowering surface tension at the air-liquid interface of the alveolus, surfactant prevents alveolar collapse at low lung volumes (Figure 1B).
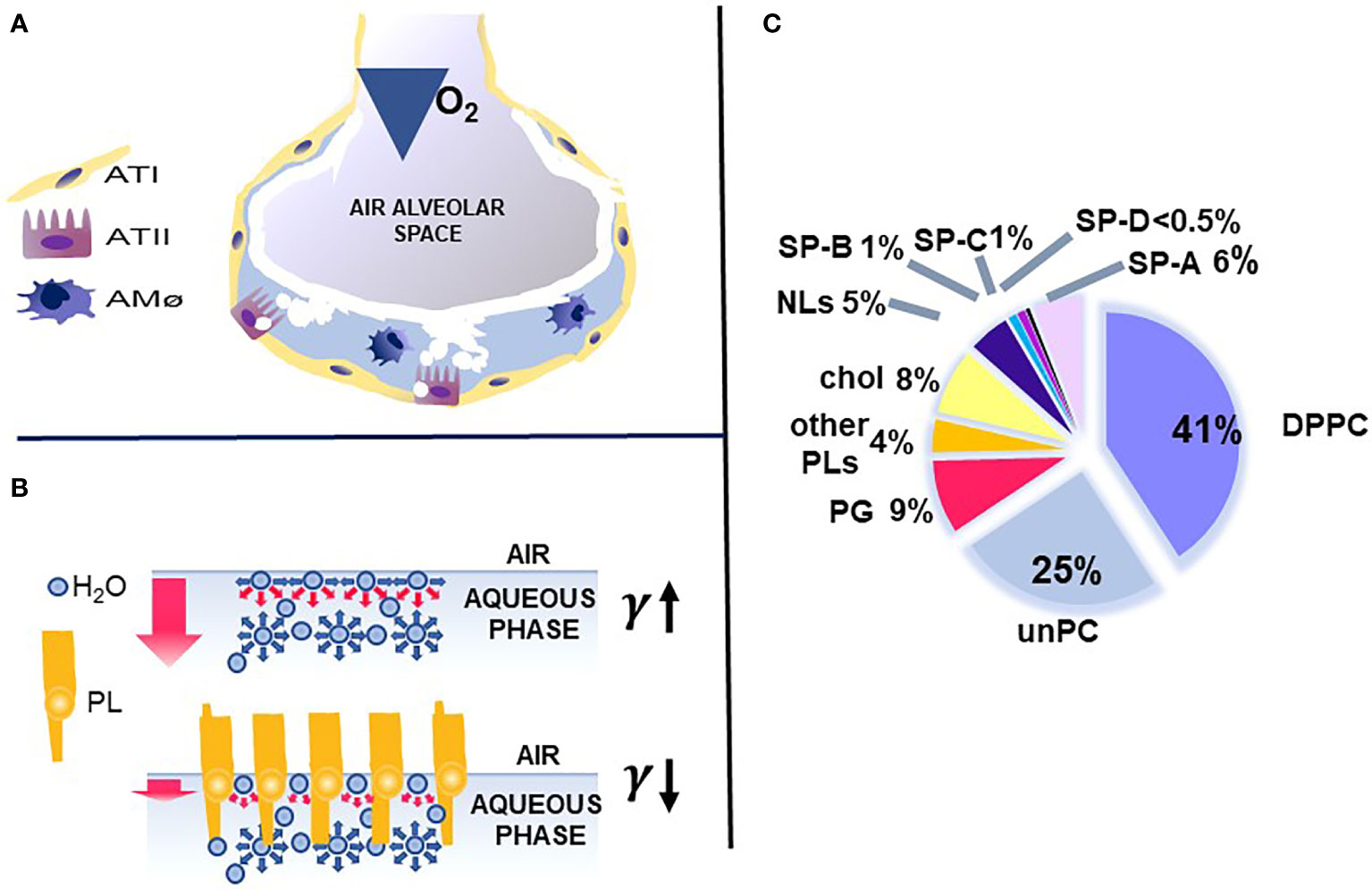
Figure 1 Alveolus, surfactant function, and surfactant composition. (A) depicts a cartoon of the alveolus, which is lined by epithelial type II and type I cells with the alveolar macrophage in the hypophase. In (B) the top air/aqueous interface lacks surfactant phospholipids and this leads to an increase in surface tension (γ). In the presence of phospholipids, the surface tension (γ) is reduced (bottom panel) and this prevents alveolar collapse. (C) depicts the lipid and protein composition of pulmonary surfactant. Courtesy of Dr. Jesus Perez-Gil (1).
Pulmonary surfactant consists of lipids, primarily phospholipids, and non-serum-derived proteins, SP-A, SP-B, and SP-C (Figure 1C). A fourth protein SP-D co-isolates with surfactant and is grouped with the other three surfactant proteins, although it is not known to play a role in any surfactant-related functions, such as surface tension reduction or surfactant structure (2) (Figure 2). In general, SP-B and SP-C, both hydrophobic proteins, are primarily involved in surfactant-related functions, and SP-A and SP-D, both hydrophilic proteins, play a role in innate immune host defense, although SP-A does contribute to various aspects of surfactant structure, and function, as depicted, in part, in Figure 2. Both SP-A and SP-D belong to the collectin family. SP-D is present on the same genetic locus as SP-A1, SP-A2 (3), and mannose-binding lectin (MBL) (4, 5), is secreted by pulmonary type II epithelial cells, and it may play a role in surfactant homeostasis (6, 7)
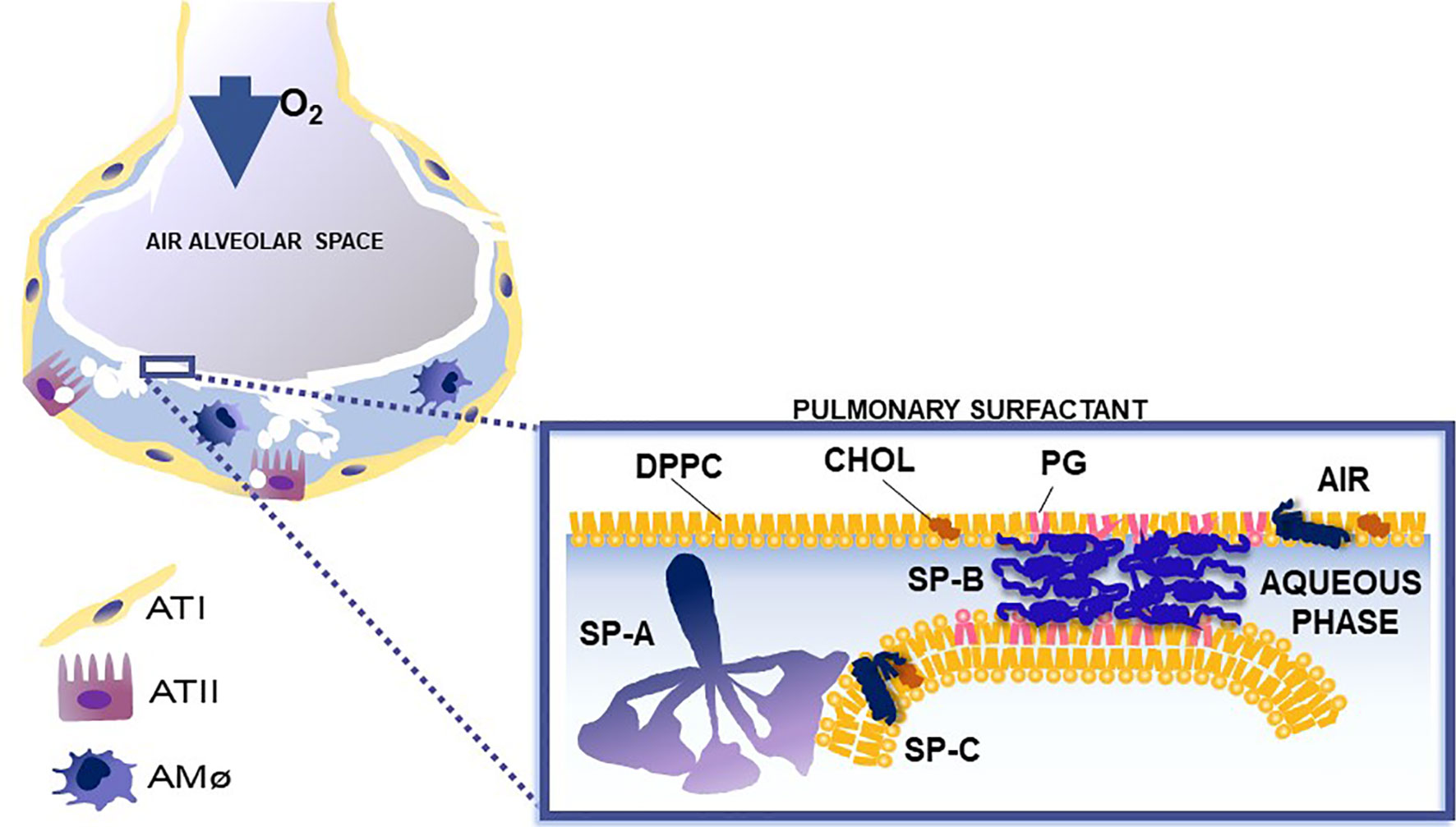
Figure 2 This cartoon depicts the role of surfactant proteins (SP-A, SP-B, and SP-C) in the biophysical properties of surfactant. Courtesy of Dr. Jesus Perez-Gil (1).
Pulmonary surfactant consists of a monomolecular surface film at the air/liquid interface of the alveolus that is directly responsible for surface tension reduction, as well as a surfactant reservoir in the hypophase (1, 8–11). In the course of a breath (compression and expansion) the surface area of the alveolus undergoes significant change. Pulmonary surfactant is reorganized and an interconnection is formed and maintained between the two associated parts of surfactant (the surface film and the reservoir). SP-B and SP-C are essential for surfactant multilayer connection and bringing lipids to the air-liquid interface. SP-A, via its ability to bind phospholipids (dipalmitoyl phosphatidylcholine) may contribute to this (12–21). SP-A1 (see below) minimizes hysteresis during the respiratory cycle and is more efficient in facilitating lipid membrane reorganization and preventing surfactant inhibition by serum proteins compared to SP-A2 (22).
The surfactant proteins, SP-B, SP-C, and SP-D, are each encoded by a single gene (2) but SP-A, the focus of this review, in humans and primates (unlike rodents) is encoded by two genes, SFTPA1 and SFTPA2 (3, 5), and their respective gene products in humans are SP-A1 and SP-A2. The two genes are distinct but share a high degree of sequence similarity (23). The human SP-A locus is mapped on chromosome 10q22.3 (24), and consists of two functional genes, SFTPA1 and SFTPA2, in opposite transcriptional orientation (3, 5). This locus also includes two pseudogenes, the MBL3P (mannose-binding lectin family member 3 pseudogene) (25) and SFTPA3P (surfactant protein A3 pseudogene (3, 5, 26). Each SP-A gene consists of four coding regions and a number of untranslated exons at the 5’ and one untranslated region at 3’ (27). A number of variants have been identified and characterized for each functional gene that may differentially affect regulation and/or function of alveolar cells, or the structure of surfactant films. These include variants resulting from alternative splicing events and variable translation initiation sites in the 5’-UTR (28–30), as well as isoforms with different signal peptide cleavage sites (23). Some of these have been shown to have an impact on translation efficiency and mRNA stability (31, 32). The 3’UTR also plays a role in SP-A regulation (33, 34). However, the differential regulation of these genes is discussed in detail in a review under preparation.
SP-A1 and SP-A2 coding variants have been identified and characterized (5, 29, 35–37). These variants are shown diagrammatically in Figure 3 and described in detail below. The cartoon in Panel B shows that both hetero-oligomers and homo-oligomers exist. Hetero-oligomers consist of two molecules or monomers of SP-A1 and one molecule of SP-A2 as described initially by Voss et al. (38) and homo-oligomers consist of three molecules of either SP-A1 or SP-A2 (23). SP-A mRNA in adult lung tissue has been localized in the epithelial type II cells by in situ hybridization (39), as has the SP-A protein by immunohistochemistry and immunoelectron microscopy (40). Although there have been published works using SP-A from various species (41–48) the focus and the reference points discussed in this review are primarily those of human SP-A1 and SP-A2. Thus, here we focus on functional and structural differences between human SP-A1 and SP-A2 and/or among their corresponding variants, as well as on their differential ability to regulate processes in alveolar cells and impact survival of the organism.
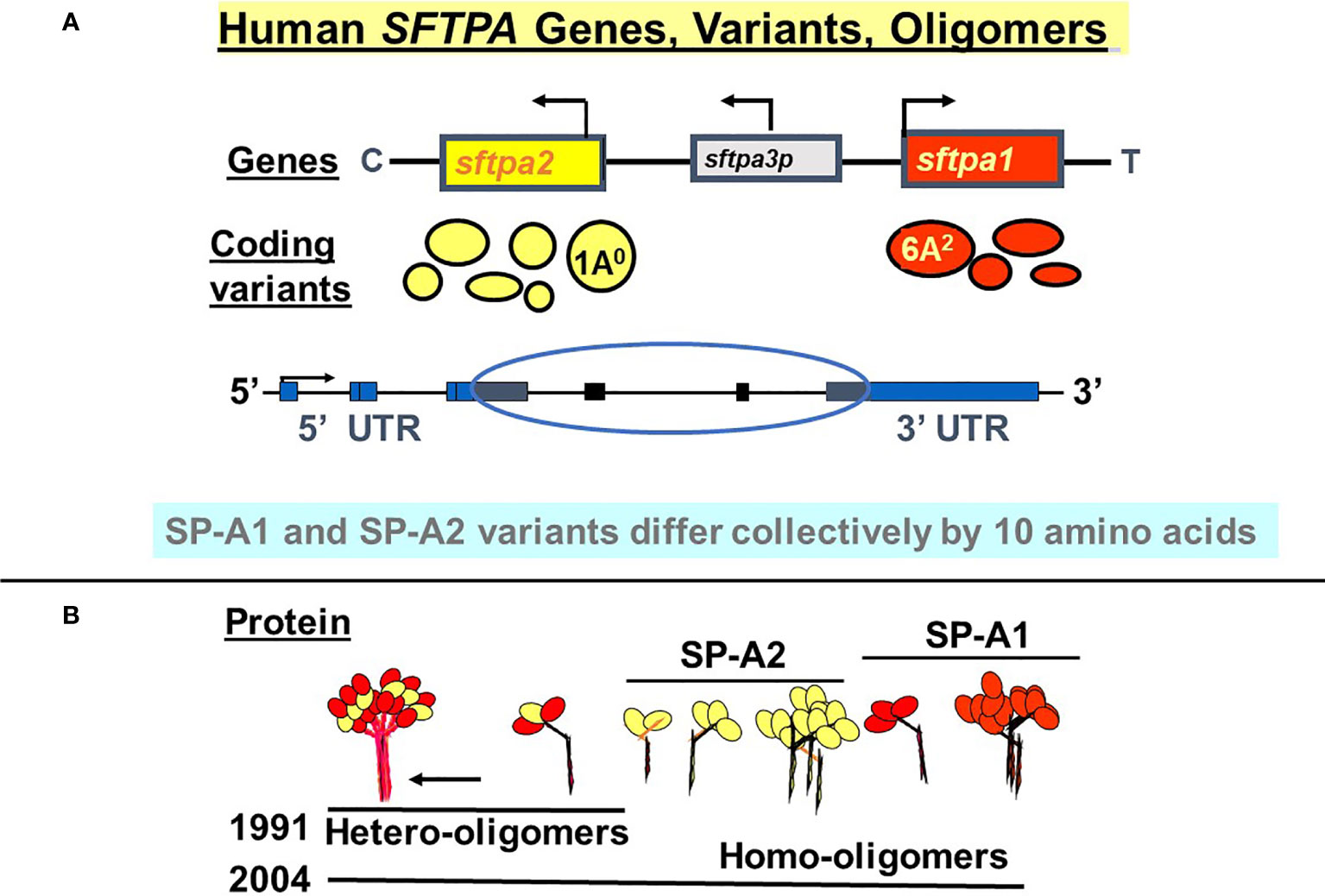
Figure 3 Human SFTPA genes, variants, and oligomers. (A) depicts the chromosomal SFTPA genetic locus from the centromere (C) to telomere (T) as well as the opposite transcriptional orientation (arrow) of the two functional genes (SFTPA1 and SFTPA2). The pseudogene (SFTPA3P) is between the two functional genes. The most frequently found coding variants are depicted below each gene. The size of the circle denotes the relative frequency with the 1A0 and 6A2 being the most frequently observed variants in the general population. A schematic of the gene organization is shown, depicting the 5’ and 3’ UTR (blue boxes) and the coding regions (black boxes, circled). (B) depicts hetero-oligomers of SP-A1 and SP-A2 described in 1991 (38) and homo-oligomers described in 2004 (23).
2. Structural Differences of SP-A1 and SP-A2 Coding Variants
The precursor molecule of each gene product contains 248 amino acids and each consists of a number of domains including a signal peptide, N-terminal region, collagen-like domain, neck, and C-terminal carbohydrate recognition domain (CRD) (5, 49–51) (Figure 4). The signal peptide of 18-20 amino acids (23) and the N-terminal region together consist of 27 amino acids, and the collagen-like domain, neck, and CRD consist of 73, 33, and 115 amino acids, respectively. The gene-specific differences that distinguish the two SP-A genes and their corresponding variants consist only of four amino acids shown in Table 1.
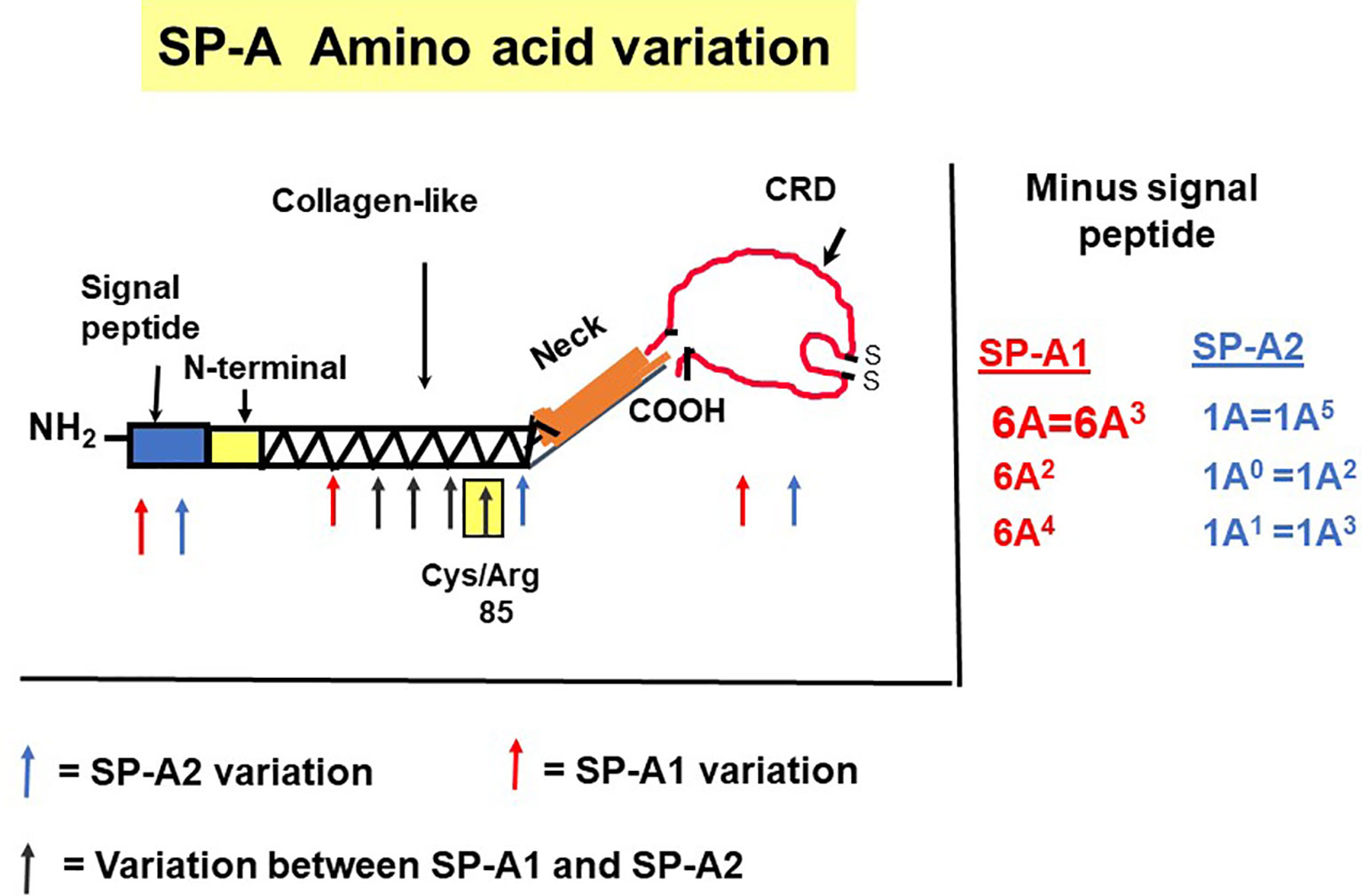
Figure 4 SP-A regions encoding the precursor molecule and the location of the gene-specific and variant-specific amino acid differences is shown (27). The numbering of the amino acids is that of the precursor molecule (i.e., includes the signal peptide). The notation of the most frequently observed variants for each gene is shown on the right. Some of the variants are identical after cleavage of the signal peptide (52).
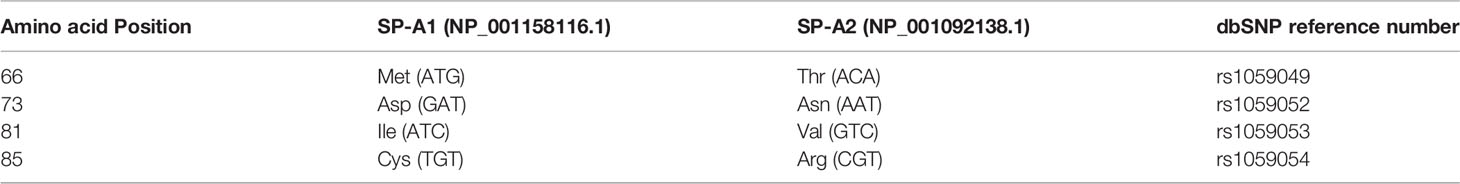
Table 1 Amino acids differences and their respective codons in parenthesis that distinguish the two human SP-A1 and SP-A2 gene products and their corresponding coding variants.
SP-A1 and SP-A2 coding variants: A number of coding variants have been identified for each SP-A gene product. The most frequently observed (>1%) in the general population are, 6A, 6A2, 6A3, 6A4 for SP-A1 and 1A, 1A0, 1A1, 1A2, 1A3, 1A5 for SP-A2 (5, 27, 49–51). These variants are found with varied frequency in the general population, with 6A2 and 1A0 being the most frequently observed (49, 50, 52, 53). The SP-A1 and SP-A2 variants, apart from the gene-specific amino acid differences shown in Table 1, differ collectively in six amino acids shown in Table 2. Gene-specific and variant-specific nucleotide differences that do not change the encoded amino acid are shown in Table 3.
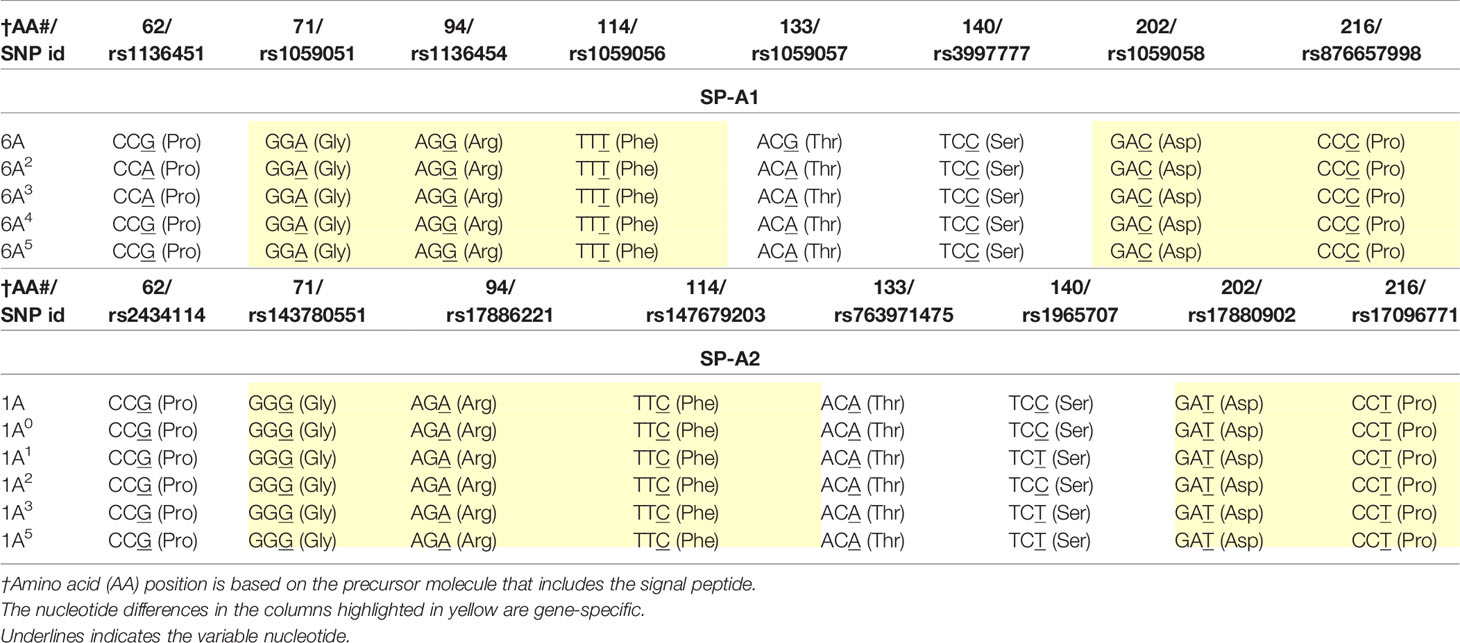
Table 3 Nucleotide differences between or among SP-A1 and SP-A2 coding variants that do not change the encoded amino acids.
All the gene-specific differences are found in the collagen-like domain of the proteins (49) (Figure 4), which consists of 23 Gly-X-Y triplets with an irregularity (Pro-Cys-Pro-Pro instead of Gly-X-Y) present between triplets 13 and 14. This irregularity generates a kink in the structure (49, 54). The four gene-specific amino acid differences at positions 66, 73, 81, 85 (Table 1) are located within the collagen-like domain at Gly-X-Y triplets, 13, 14, 17, and 18, respectively. Of the gene-specific amino acid differences, amino acid 85 is shown to affect both structure and function of the SP-A protein. When mammalian (CHO) cell-expressed wild type (WT) and mutated proteins containing interchanged residues in position 85 (SP-A1C85R and SP-A2R85C) were compared, the oligomerization patterns were largely determined by the amino acid at position 85 (55). These results were consistent with, and confirmed oligomerization patterns observed, in previous studies (23). In humanized transgenic mouse models where each mouse carries and expresses a different SP-A1 or SP-A2 variant (56), SP-A1 forms dimers, hexamers, and larger oligomer complexes under non-reducing conditions, while SP-A2 forms mostly dimers, trimers, tetramers, and hexamers (56). Differences in oligomerization were also observed in SP-A1 and SP-A2 proteins expressed in insect cells (57), although the pattern differed from that in mammalian cell-expressed SP-As. SP-A undergoes significant co- and post-translational modifications, some of which cannot be performed by insect cells. For example, insect-cell expressed SP-A is deficient in hydroxyproline (58) and hydroxylation of prolines retards the electrophoretic mobility of SP-A (59) as well as increases the melting temperature (Tm) compared to molecules lacking proline hydroxylation (60). Insect cell-expressed SP-A2 demonstrated higher rough LPS aggregation activity than SP-A1 (54, 61). SP-A2 also exhibited a higher activity to self-aggregate and induce aggregation of phospholipids and bacterial lipopolysaccharides than co-expressed or co-ex (SP-A containing both SP-A1 and SP-A2) or SP-A1 proteins (SP-A2 > SP-A1/SP-A2 > SP-A1). This indicates that SP-A1, either in homo-oligomers (i.e., by itself) or hetero-oligomers (i.e., in the presence of SP-A2), exhibits a lower ability to self-aggregate (54). Moreover, SP-A2 was shown, via circular dichroism and fluorescence studies, to be more stable than SP-A1 (54). Based on substitution experiments of amino acids X and Y in Gly-X-Y triplets in other systems (62, 63), it was postulated that the presence of Cys85 (in SP-A1) within the collagen-like domain creates a micro instability contributing, perhaps, to a lower SP-A1 stability compared to SP-A2 (Arg85) (54).
In vitro experiments of mammalian expressed proteins showed that the order of the proteins with regards to their activity in aggregating rough LPS was SP-A2WT>SP-A1WT>SP-A1C85R>SP-A2R85C (55). Substitutions in residue 85 affected LPS aggregation as well as phagocytosis of P. aeruginosa by male rat alveolar macrophages (AM). The phagocytic index of mutated and wild type (WT) SP-A1 and SP-A2 was SP-A1C85R > SP-A2WT > SP-A1WT = SP-A2R85C (55). Cysteine 85 of SP-A1 allows formation of additional intermolecular disulphide bridges that affects oligomerization patterns (23). Furthermore, introduction of cysteine 85 in SP-A2 changes its oligomerization pattern to one that is identical to SP-A1, while introduction of arginine 85 in SP-A1 increased the efficiency of rat AMs to phagocytose bacteria (55). Importantly, the natural presence of arginine 85 in SP-A2 introduces an additional trypsin digestion site, which makes the protein more susceptible to degradation by trypsin at 37°C (61). These results collectively highlight the importance of residue 85. In addition to the importance of residue 85, the presence of Trp (instead of Arg) in position 219 of the SP-A1 6A2 variant, which is associated with increased risk for idiopathic pulmonary fibrosis, has been shown to mediate differences in protein self-aggregation before and after ozone exposure (64).
3. Surfactant Structure- and Function-Related Differences in the Presence of SP-A1 and SP-A2
The importance of the SP-A collagen-like domain in surfactant-related function/structure was highlighted in transgenic mice carrying a truncated form of SP-A with a deletion of amino acids 6-80 on the SP-A-/- (knockout) background (65). These mice could not form tubular myelin, an extracellular structural form of surfactant, which is considered to be a surfactant reservoir. The mouse SP-A has a histidine in position 85 and the rat, an arginine. Both of these residues are basic polar and thus similar to SP-A2, which contains an arginine at position 85. The SP-A1, on the other hand, has a cysteine at position 85 and this could mediate intra- and inter-molecular disulfide bonds that none of the amino acids at position 85 in rodent SP-A or SP-A2 could form (23). An amino acid alignment of all SP-A species sequences available along with their sequence similarity is depicted in Figure 5A. Given the importance and magnitude of the impact of residue 85 on structure and function (based on experimental evidence), a few comments are warranted about this residue in the various species. Humans and primates have two SFTPA genes whereas all other mammalian species known to date have a single SFTPA gene. The SFTPA1 gene in humans and primates (chimpanzee) that encodes SP-A1 has a Cys at residue 85 but none of the other sequences has a Cys at this residue. The second SFTPA gene in humans encodes SP-A2 and has an Arg at residue 85. Similarly, all of the other sequences have an Arg except for the mouse sequence that has a histidine and the primate SFTPA2 that has a Leu, making all these sequences, based on residue 85, similar to SP-A2. Panel B of Figure 5 depicts a phylogenetic tree of the species shown in the alignment in Panel A of Figure 5. Amino acid differences in other residues have not been functionally examined via amino acid substitution experiments as residue 85 has. However, although these amino acids may contribute to various SP-A-mediated functions these may not impart the magnitude of functional differences caused by residue 85 (55). Interestingly, a recombinant trimeric fragment of human SP-A1 that contains the CRD, the neck, and the last 8 Gly-X-Y repeats (including Cys85) is efficacious at neutralizing respiratory syncytial virus and preventing infection of differentiated human bronchial epithelial cells (66).
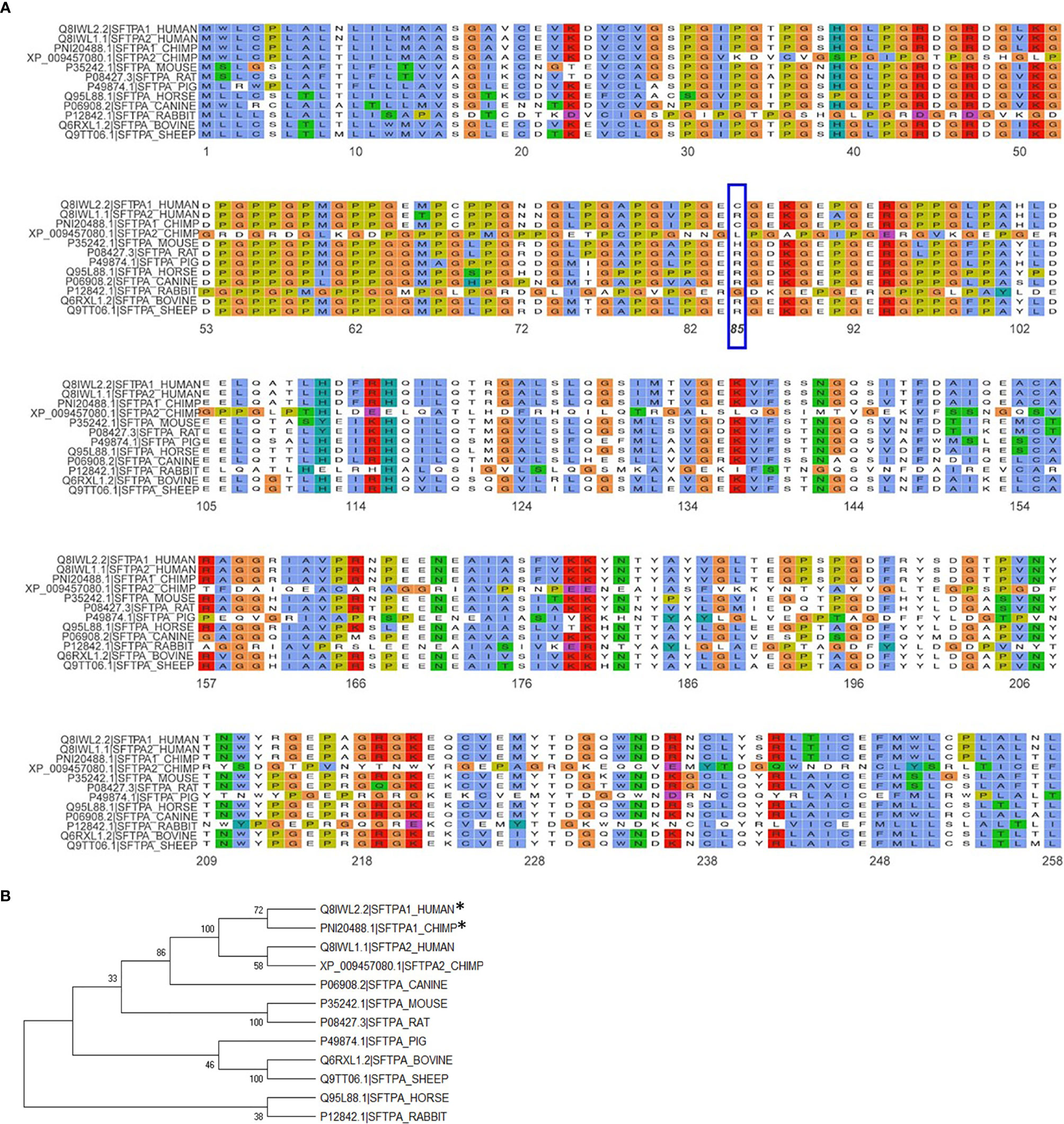
Figure 5 Alignment and phylogeny of SP-A1 and SP-A2 sequences. (A) shows the alignment of the NCBI available SP-A sequences for the species shown using the ggmsa R package (http://yulab-smu.top/ggmsa/index.html). Per NCBI statement “this record is predicted by automated computational analysis. This record is derived from a genomic sequence (NC_036889.1) annotated using gene prediction method: Gnomon, supported by mRNA and EST evidence”. Gnomon, a gene prediction tool, looks for ORFs, splice sites etc. The amino acid of residue 85 is highlighted by the blue box. The color scheme follows the default colors used in alignments in Clustal X. (B) shows the phylogenetic tree of the sequences shown in (A). The tree was created with the MEGA X software (www.megasoftware.com), using the neighbor-joining method. The numbers next to the branches represent the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The asterisk mark the two sequences, human SP-A1 and primate SP-A1 sequence, that have a C (Cys) at residue 85. All other species sequences have an R (Arg) except for the mouse that has a H (His) and the primate SFTPA2 gene that has an L (Leu), as shown in (A).
Tubular myelin, an extracellular structural form of surfactant, was not observed in hTG mice expressing either SP-A1 or SP-A2, although both types of mice had lamellar bodies, and SP-A1-expressing mice had larger lamellar bodies (56). Tubular myelin was, however, present in SP-A1/SP-A2 co-expressing transgenic mice, indicating the need for both gene products for tubular myelin formation (56). Moreover, a rescue study of SP-A knockout (KO) mice with exogenous human SP-A from bronchoalveolar lavage (BAL) showed that tubular myelin was restored, when the exogenously administered SP-A contained both SP-A1 and SP-A2. But this was not the case if the exogenous SP-A contained primarily SP-A1 or SP-A2 (56) further supporting the observation that both SP-A1 and SP-A2 gene products are necessary for tubular myelin formation. Furthermore, SP-A has been localized in tubular myelin using a number of methods (40, 67).
A study using a captive bubble surfactometer to investigate the biophysical properties of lung surfactant from humanized transgenic mice expressing either protein or both SP-A proteins showed that the presence of SP-A1 minimizes hysteresis (22). Hysteresis has been described and studied extensively elsewhere by experts in the field of pulmonary mechanics and in biophysical studies of pulmonary surfactant (68–71). In the study by Lopez-Rodriquez (22) SP-A1 proved to be better in enabling lipid membrane reorganization by more efficiently bringing together membrane aggregates to form complex films (Figure 6) and showed a higher efficiency in phospholipid adsorption at the air-liquid interface. Moreover, when experiments were performed at surfactant concentrations to detect differences between SP-A1 and SP-A2, SP-A1-containing preparations lowered surface tension more efficiently, even in the presence of serum (22), indicating that SP-A1 has a better ability to prevent inhibition of surfactant function by serum proteins.
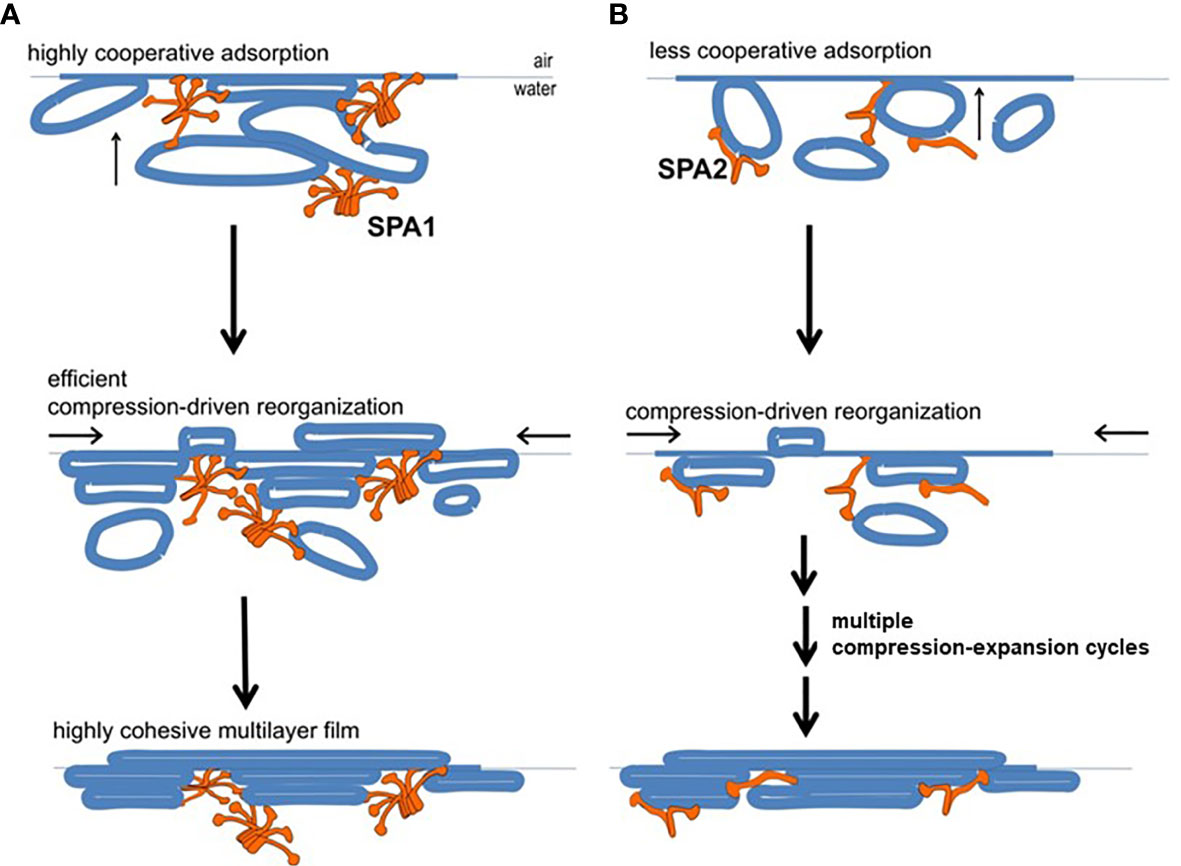
Figure 6 Model of behavior of surfactant lipid complexes in the presence of SP-A1 or SP-A2 proteins. (A) depicts the multivalent binding capacities of SP-A1. The higher SP-A1 multivalent binding capacity is advantageous as it brings together several membrane aggregates and forms complex films. This is due to its higher oligomerization state that is mediated in part by the presence of Cys85. These complex films, upon compression-driven reorganization, could rapidly generate highly cohesive multilayer films that can reach very low surface tension. (B) depicts how SP-A2, which is less oligomerized and exhibits lower binding capacity, forms surface films with lower density of membranes (i.e., less complex films). These simpler films can still be reorganized in a cohesive multilayer film to impart low surface tension but they require repetitive compression-expansion cycling. Therefore, part of the energy needed for the initial compression-expansion cycles will be spent to promote folds and membrane-membrane contacts needed prior to generating the cohesive multilayer film, making this process less efficient (22).
Moreover, SP-A2 is more effective than SP-A1 in promoting dipalmitoyl phosphatidylcholine (DPPC) and dipalmitoyl phosphatidylglycerol (DPPG) lipid vesicle aggregation (54, 61). DPPC monolayers containing porcine SP-B, in the presence of SP-A2, are morphologically closer to the ones containing native SP-A (i.e., SP-A from BAL). This is not the case with SP-A1, where although SP-A1 allows the formation of SP-B/DPPC monolayers, these differ compared to those with native SP-A (72). However, the lipid SP-B containing films in the presence of both SP-A1 and SP-A2 (i.e., co-ex proteins) showed combined characteristics of films containing either SP-A1 or SP-A2. It is possible that varying levels of SP-A1 and/or SP-A2 (73) in BAL may differentially affect the physiological properties of surfactant and be critical for lung health.
4. SP-A1 and SP-A2 Differentially Affect the Activity of Alveolar Macrophages (AMs) Ex-Vivo
The sentinel innate immune cell in the alveolus is the AM, and SP-A is shown to interact with the AM (74–77). SP-A1 and SP-A2 variants differentially affect several of the processes and functions of AM, as discussed below. SP-A2 enhances bacteria phagocytosis by AM more than SP-A1. Insect-cell expressed SP-A2 was more active than SP-A1 in its ability to increase association of rat AM with Pseudomonas aeruginosa in all concentrations studied (78). Although the observations from CHO-cell expressed SP-A1 and SP-A2 proteins (79) with regards to bacterial phagocytosis were similar to those observed by insect-cell expressed SP-As, the concentration of the CHO-cell expressed protein needed for this effect was orders of magnitude lower (79). SP-A undergoes a significant number of co- and post-translational modifications (59, 80–84), but as mentioned earlier, the insect-cell derived SP-A proteins lack certain post-translational modifications (58). The significantly higher concentration of insect-expressed protein needed for a similar functional activity as that observed with CHO-derived SP-A protein (79) indicates that the co- and post-translational modifications of SP-A are necessary for optimal function.
As discussed above, a number of SP-A1 and SP-A2 variants have been identified and the variants of each protein studied differentially impact AM function and thus are of importance. Of the SP-A2 variants studied, in terms of their ability to enhance bacterial phagocytosis, the 1A1 variant was more efficient than 1A0, and of the SP-A1 variants, the 6A2 was more efficient than 6A4. Apart from the SP-A1 and SP-A2 gene-specific differences shown in Table 1, the SP-A2 variants 1A1 and 1A0 differ in three residues, at positions 9, 140, and 223 (Tables 2 and 3), while the SP-A1 variants differ in three residues, at positions 50, 62, and 219 (Tables 2 and 3) (27, 49, 53). While the degree to which these amino acids contribute to various functions is not entirely known, the amino acid differences of these SP-A1 and SP-A2 variants collectively are located within the collagen-like domain (amino acid 50) and the CRD (amino acids 219 & 223) of the mature protein. Nucleotide polymorphisms at residues 62 and 140 do not change the encoded amino acid and residue 9 is part of the signal peptide (27). However, the gene-specific difference Cys/Arg at residue 85 in SP-A1 and SP-A2, respectively, has a significant effect on AM function including on bacterial phagocytosis (55), perhaps partly due to the structural changes this residue imparts on SP-A1 and SP-A2 (i.e., oligomerization, stability, other). As noted, the contribution of amino acids other than Arg/Cys85 in functional and/or structural differences has not been studied in detail. We speculate that although the magnitude of change of Arg/Cys85 on function/structure is significant, the amino acid differences in other SP-A1 and SP-A2 residues serve as a rheostat to modulate the overall function of each SP-A1 or SP-A2 variant. This postulate is consistent with findings in several functional read-outs, including bacterial phagocytosis discussed above and others discussed below. As a group, SP-A2 variants exhibited higher activity than SP-A1 variants in their enhancement of the bacterial phagocytic index (78, 79, 85), but the variants within each group, expressed either in mammalian CHO cells (79) or insect cells (78), also showed differences in their effects on the phagocytic index. Whether the higher SP-A2 activity is due, in part, to its CRD being able to bind a wider variety of sugars (on pathogen surfaces) compared to SP-A1 (86) remains to be determined. Comparable observations were made on SP-A1 and SP-A2 variants in their ability to enhance pro-inflammatory cytokine production by a macrophage-like cell line (57, 87, 88).
Exposure of SP-A to ozone (O3) (89) decreases its ability to interact with alveolar macrophages (90, 91). Following oxidation of CHO-derived SP-A, the ability of SP-A2 to enhance phagocytosis of P. aeruginosa by rat alveolar macrophages remained higher than SP-A1, albeit, compared to SP-A1, the drop in activity before and after O3 exposure was larger in SP-A2, indicating that SP-A2 is more sensitive to O3 exposure than SP-A1 (85). Similarly, ozone exposure of SP-A variants had a negative impact on SP-A-regulated cytokine production (57, 88).
Other: Despite their differential impact on macrophage activity, the two SP-A proteins did not seem to differ in their activity against the influenza A virus (92). This, however, may need to be studied further because the SP-A proteins studied were expressed in CHO cells. These cells lack α(2,6)-sialyltransferase (93, 94) and thus they cannot synthesize α(2,6)-linked sialic acids in the expressed proteins, but they do form α(2,3)-linked sialic acids (92). Insect-cell expressed SP-As, on the other hand, that lacked sialic linkages altogether failed to inhibit the hemagglutination activity of influenza A virus (92). The presence of the specific sialic acid linkages for appropriate SP-A function may be critical in response to influenza A virus. These linkages or other co-/post-translational modifications or genetic variations may in fact differentially contribute to SP-A1 and SP-A2 variant function in response to influenza A virus infection as suggested by human association studies (95). Furthermore, an SP-A2 223Q variant (glutamine residue at position 223) was shown in mice to provide significant protection from ozone-induced airway hyper-responsiveness compared to another SP-A2 223K variant (lysine at 223) and the SP-A KO mice (96).
5. Differences in the Bronchoalveolar Lavage (BAL) and/or Alveolar Macrophage (AM) Proteome of Various SP-A Mouse Models and Conditions
5a. SP-A Knockout (KO) and Wild Type (WT) Mice
BAL proteome: The mouse BAL proteome is affected by the presence of SP-A. The proteins mostly affected at basal conditions in the KO vs WT are involved in host defense, redox equilibrium, and protein modifications (97). Proteins with a role in host defense against Klebsiella pneumoniae (98, 99), were elevated in the SP-A KO mouse under basal conditions, indicating potential compensatory mechanisms for host defense deficits in the KO. However, these do not seem to be adequate in the long term, because SP-A-KO mice infected with K. pneumoniae are shown to exhibit significantly reduced survival compared to WT mice (100). Moreover, the levels of these proteins (lysozyme and β2-microglobulin) were decreased in infected KO and the levels of other proteins involved in host defense and redox balance were altered in infected KO compare to WT infected mice. These together indicate that in the absence of SP-A there may be a loss of control to effectively regulate expression of such proteins in response to infection. Furthermore, lipopolysaccharide (LPS) treatment of KO mice negatively impacts markers of inflammation long after treatment and the KO become less capable of responding to a second insult (101).
Similar biological pathways were affected in the O3-challenged KO mouse model. The response pattern of the BAL proteome of KO mice exposed to O3 vs filtered air-exposed, is magnified compared to that of similarly treated WT mice (102). An interesting observation from the same work was the similarities in the levels of changed proteins observed between filtered air-exposed KO mice and O3-exposed WT mice, indicating that the absence of SP-A may place the alveolar cells in a state of chronic oxidative stress, as may occur after O3 exposure in WT mice. Moreover, the magnitude of several protein changes in response to infection or O3 exposure in the KO and WT differed, with the KO exhibiting a higher degree of response. A possible explanation would be that SP-A plays a role in processes involved in infection and/or oxidative stress and its absence generates a deficit in host defense functions and/or loss of regulatory control leading to an overexuberant response in KO in the face of insult. A recent study, using optical redox imaging showed that AM from KO are more oxidized after O3 exposure compared to AM that were chronically or constitutively exposed to SP-A2 (103).
AM proteome: Because SP-A has been shown to modulate several AM functions, it was postulated that several of the protein changes found in the KO and WT BAL proteome are partly due to changes in the AM proteome. A rescue proteomics experiment where SP-A KO mice were treated with a single dose of human SP-A, revealed that the proteome of rescued AM, resembled that of wild type (WT) AMs (104, 105). The fact that a single exogenous dose of SP-A is sufficient to nearly restore the male AM proteome to its WT state (104, 105) provides support for a role of SP-A in the regulation of the AM proteome and consequently the BAL proteome. More importantly, this highlights the potential of SP-A therapy. Sex differences were also observed in this rescue study (104, 105). A number of proteins, such as major vault protein, chaperonin subunit (beta) (CCT2), and Rho GDPα dissociation inhibitor, found to be increased in females, are shown to interact with the estrogen receptor (106–108) indicating that hormones may play a role in mediating SP-A action. Consistent with this notion is the finding that: a) gonadectomy eliminated (females) or significantly reduced (males) survival differences of infected mice in the presence or absence of O3-exposure; and b) sex hormone replacement in gonadectomized mice had a significant impact on the survival of the male and female mice after K. pneumoniae infection (109). The patterns of survival after hormone replacement of the gonadectomized mice resembled those of intact mice (109). In summary, SP-A appears to regulate hormone-dependent genes/processes. Although the mechanisms involved are not understood yet, this is an important topic and warrants further investigation.
Differences in specific protein levels were observed in AM in the presence or absence of SP-A. These proteins were involved in several pathways including pathways pertaining to actin cytoskeleton signaling, Nrf2-mediated oxidative stress, regulation of inflammation, protease/chaperone function, cell differentiation and other (Figure 7A) (105). A single dose of SP-A treatment of AM in culture obtained from SP-A KO mice, significantly decreased F-actin intensity/pixel as early as one-hour post treatment (Figure 7B) and increased AM cell size (Figure 7C) (105, 110), indicating that in response to SP-A a number of cytoskeletal events may occur. These may include a redistribution of cortical actin over a larger cell area and/or altered gene expression of actin-related proteins (105). These changes may underlie functional activities of the AM, such as bacterial phagocytosis, observed at one hour after AM are incubated with SP-A and bacteria (78, 79, 85).
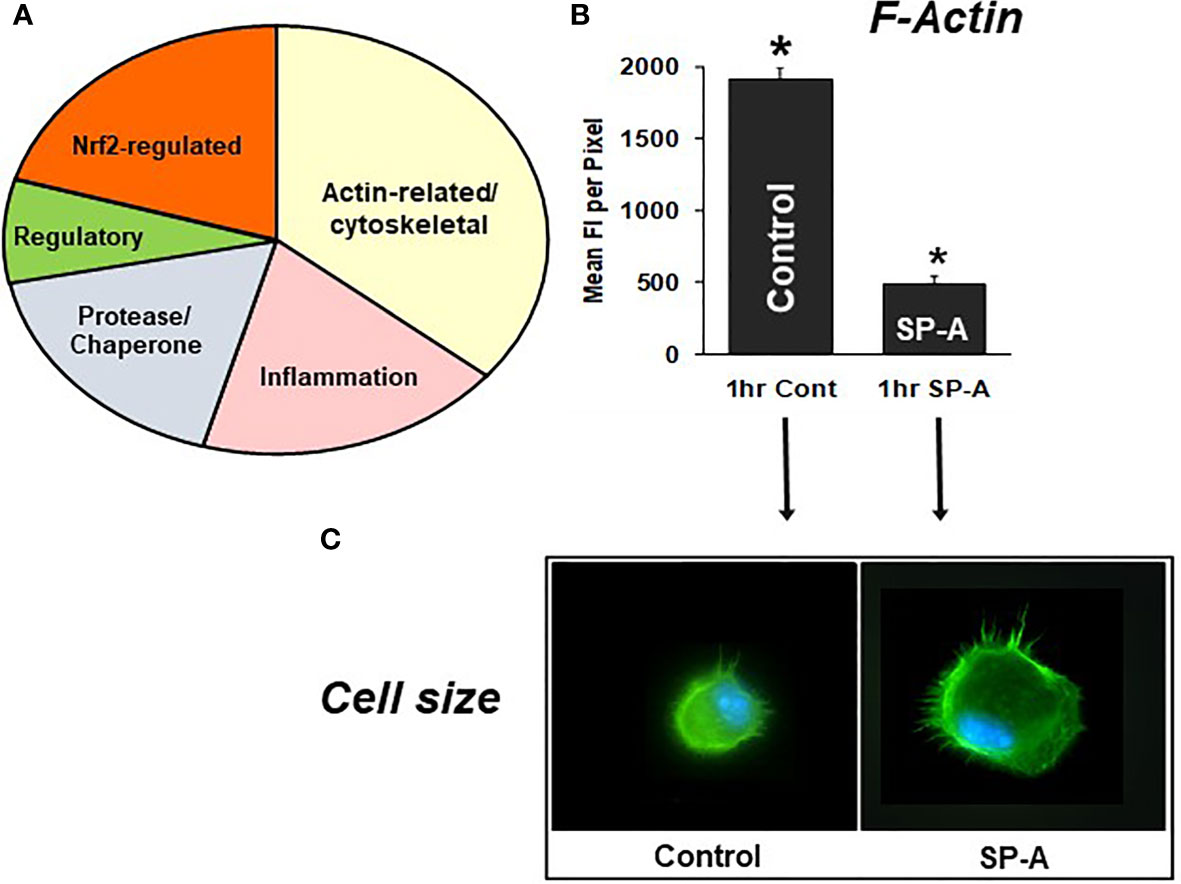
Figure 7 SP-A rescue of the SP-A KO AM. (A) depicts the groups of proteins that changed in response to SP-A. (B) depicts the mean fluorescence per pixel of F-actin (cells stained with Alexa Fluor 488 phalloidin) after 1hr of treatment with SP-A (1hr SP-A) or untreated (1hr control) SP-A KO AM (105). (C) depicts the actual fluorescence in the control and SP-A-treated AM as well as the size change after the treatment (110). AMs were visualized using a Nikon TE-2000 PFS fluorescent microscope with a 60X (1.4 NA) phase contrast lens with a 1.5× tube lens in place and the images captured using a Photometrics Coolsnap HQ2 digital camera (0.07 μm/pixel) and saved as TIFF files (105). *p < 0.0001.
5b. Humanized Transgenic (hTG) Mice Where Each Carries and Expresses a Single SP-A1 or SP-A2 Variant, or Both
BAL proteome: The BAL proteomes of SP-A1 and SP-A2 hTG mice are differentially affected by human SP-A variants (111). In terms of the number of proteins that changed (more than 25% in BAL), the male SP-A2 mice exhibited a more robust response (SP-A2 > SP-A1 = KO) following K. pneumoniae infection in the presence of O3 exposure (111). Moreover, the majority of the changed proteins showed increases in males and decreases in females. However, in the absence of O3 exposure following infection the SP-A1 and SP-A2 variants differentially affected the BAL proteome in males (SP-A2 > SP-A1 > KO), but less so in females (SP-A2 = SP-A1 > KO) (111). The top pathways implicated in these differences are the acute phase response (APR) that includes the NF-kB signaling pathway, and the Nrf2-mediated oxidative response, among others. A time-dependent Nrf2 translocation from the cytoplasm to the nucleus was observed after O3 exposure (111). Nrf2-regulated proteins were also identified in the proteome of the AM in an SP-A rescue experiment of SP-A KO mice (104), further supporting a role of SP-A in this signaling pathway. Compared to KO, the effect of SP-A2 on signaling pathways, such as APR and Nrf2, exhibited sex differences and in males appeared to be independent of O3-exposure, whereas this was not entirely the case in SP-A2 females, especially for Nrf2. Under similar conditions SP-A1 exhibited sex differences both in response to O3 and filtered air exposures. The SP-A1 effect on Nrf2 was eliminated after O3 exposure in both males and females. These findings indicate a higher ability of SP-A2 to elicit innate immune responses, with sex differences being observed with SP-A1 and SP-A2.
AM Proteome: The proteome profile of AMs from humanized transgenic mice that express human SP-A1 or SP-A2 showed dose and sex-dependent differences (112). The SP-A2 (but not the SP-A1) profile shared similarities with that of WT, especially in proteins related to actin cytoskeleton and Nrf2-regulated proteins. However, the expression pattern of AM-derived from hTG mice expressing high levels of SP-A1 or SP-A2 was the inverse for many proteins, i.e., proteins with high levels in one type of hTG mouse (i.e., SP-A1 or SP-A2) were found with low levels in the other and vice versa. This is in contrast to the pattern observed in AM derived from hTG mice expressing low levels of SP-A1 or SP-A2 (112) where the response pattern of the two proteomes was similar. One possibility is that low levels of SP-A1 or SP-A2 affect the AM via a high affinity non-SP-A variant-dependent receptor, whereas at high levels, SP-A1 or SP-A2 may act via a different receptor or via the same one, but with different binding capacities resulting in response differences. Consistent with this, SP-A2 is shown to exhibit a higher binding capacity to AM compared to SP-A1. Moreover, SP-A1, in contrast to SP-A2, showed no binding to AMs that were not previously exposed to SP-A (i.e., in KO AMs) (113). Collectively, these indicate potential complexities of SP-A interactions with AMs. SP-A1 may have an inhibitory effect on AM binding by SP-A2 since the SP-A2 binding capacity was reduced when AMs were exposed continually to SP-A1 prior to the assay (113).
A rescue proteomic experiment of SP-A KO mice with a single dose of either SP-A1 or SP-A2 showed variant-specific protein changes as well as sex-specific changes for various groups of proteins (114). The differentially expressed proteins were grouped in one of the following categories: protease balance/chaperone function group, actin-related cytoskeleton, Nrf2-related proteins, regulatory/differentiation-related proteins, and regulation of inflammation-related proteins (114). Responses to the different SP-A gene products of these groups of proteins were sex-specific. Females were more responsive to SP-A1 with respect to proteins involved in actin metabolism and oxidative stress, and males were more responsive to SP-A2 treatment, particularly with regards to the regulation of proteins involved in protease balance/chaperone function and in inflammation (Figure 8). The functional group that was similar between males and females exposed either to SP-A1 or SP-A2 was the one containing regulatory/developmental-related proteins (114). Of interest, the response pattern of most of the proteins involved in “the regulation of actin-based motility by Rho” pathway in male and female AM rescued by SP-A1, was the inverse of one another (114).
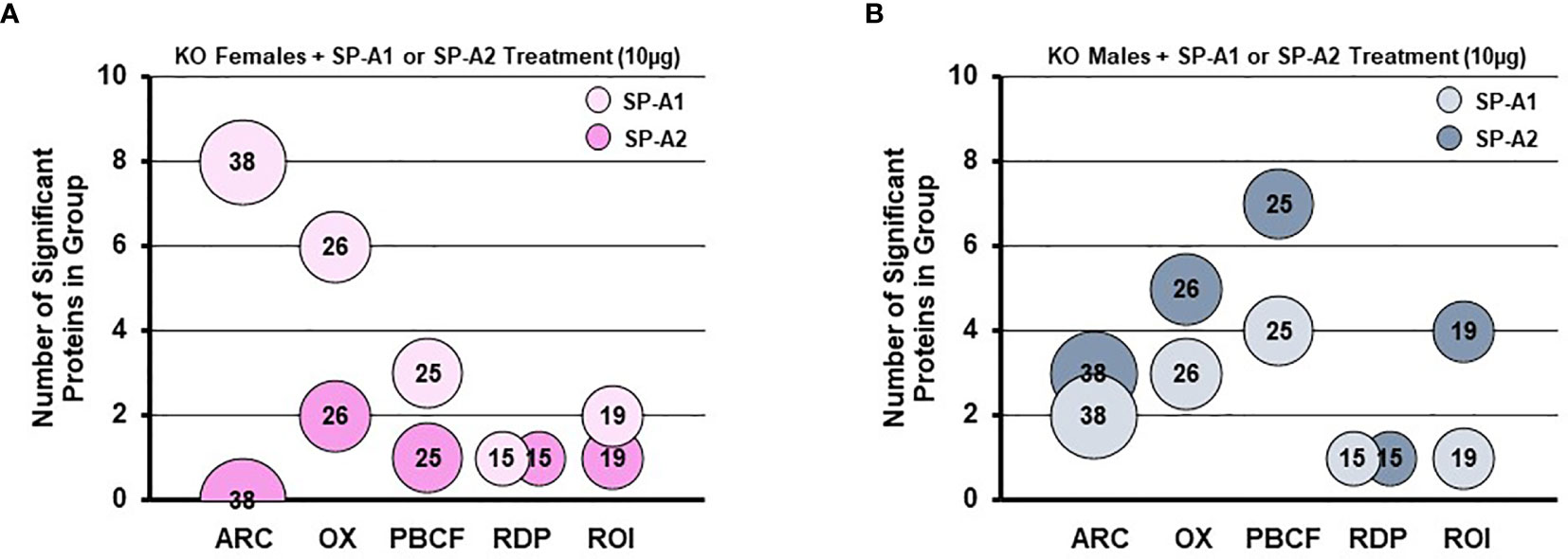
Figure 8 Sex differences of the AM proteome in response to in vivo rescue of SP-A KO mice with different human SP-A variants. (A, B) show differential effects of SP-A1 and SP-A2 in females (A) and males (B) in terms of various functional protein groups. Bubble size and number depict the number of proteins within each group (114). ARC, actin-related cytoskeletal; OX, oxidative stress; PBCF, protease balance/chaperone function; RDP, regulatory/developmental processes; ROI, regulation of inflammation.
The BAL and AM proteomics data collectively indicate that: a) SP-A plays an important role in the expression of proteins found in bronchoalveolar lavage (BAL) and those of the AM, under baseline conditions and in response to various insults, such as infection and oxidative stress. b) Sex-specific and SP-A variant-specific changes exist. c) An SP-A dose effect exists, where the proteome profile of AM from mice expressing high levels SP-A1 and SP-A2 differed, whereas the AM proteome from mice expressing low levels showed no major differences. d) Several pathways were identified in response to SP-A and between SP-A1 and SP-A2 variants under various conditions that included the actin cytoskeleton, Nrf2, NF-kB, and other. This complex interplay of SP-A variants and their total protein levels, sex, and exposure (whether environmental or infectious) that together lead to varying outcomes under the studied conditions, point to opportunities for further research into underlying mechanisms. Understanding mechanisms of SP-A variant-mediated regulation of sex-dependent molecules/processes may help in personalized medicine decisions when the use of SP-A therapy may be considered.
6. Differential Impact of SP-A1 and SP-A2 on Cytoskeleton, miRNome, Gene Expression and Toponome of Alveolar Cells
6a. Actin Cytoskeleton
SP-A has been shown to induce changes in the actin cytoskeleton of AMs (115). AMs from SP-A1 (6A2) and SP-A2 (1A0) hTG mice showed via proteomic analysis, among others, significant differences in heat maps of actin-related/cytoskeletal proteins (112), pointing to differences in actin cytoskeletal processes. Figure 9 shows that the pattern of F-actin fluorescence distribution differs between SP-A1 and SP-A2 AM stained with AlexaFluor phalloidin and that F-actin is decreased in SP-A2 compared to SP-A1 AM, as assessed by measurements of the fluorescence intensity/pixel (Panel A) or by Western blot (Panel B) (110). Moreover, the ARP3 that regulates actin polymerization is also decreased in SP-A2 AM (Figure 9C), consistent with the lower F-actin levels (110). We postulate that the state of the cytoskeleton in SP-A2 AM makes the AM more agile for movement and function (i.e., bacterial phagocytosis).
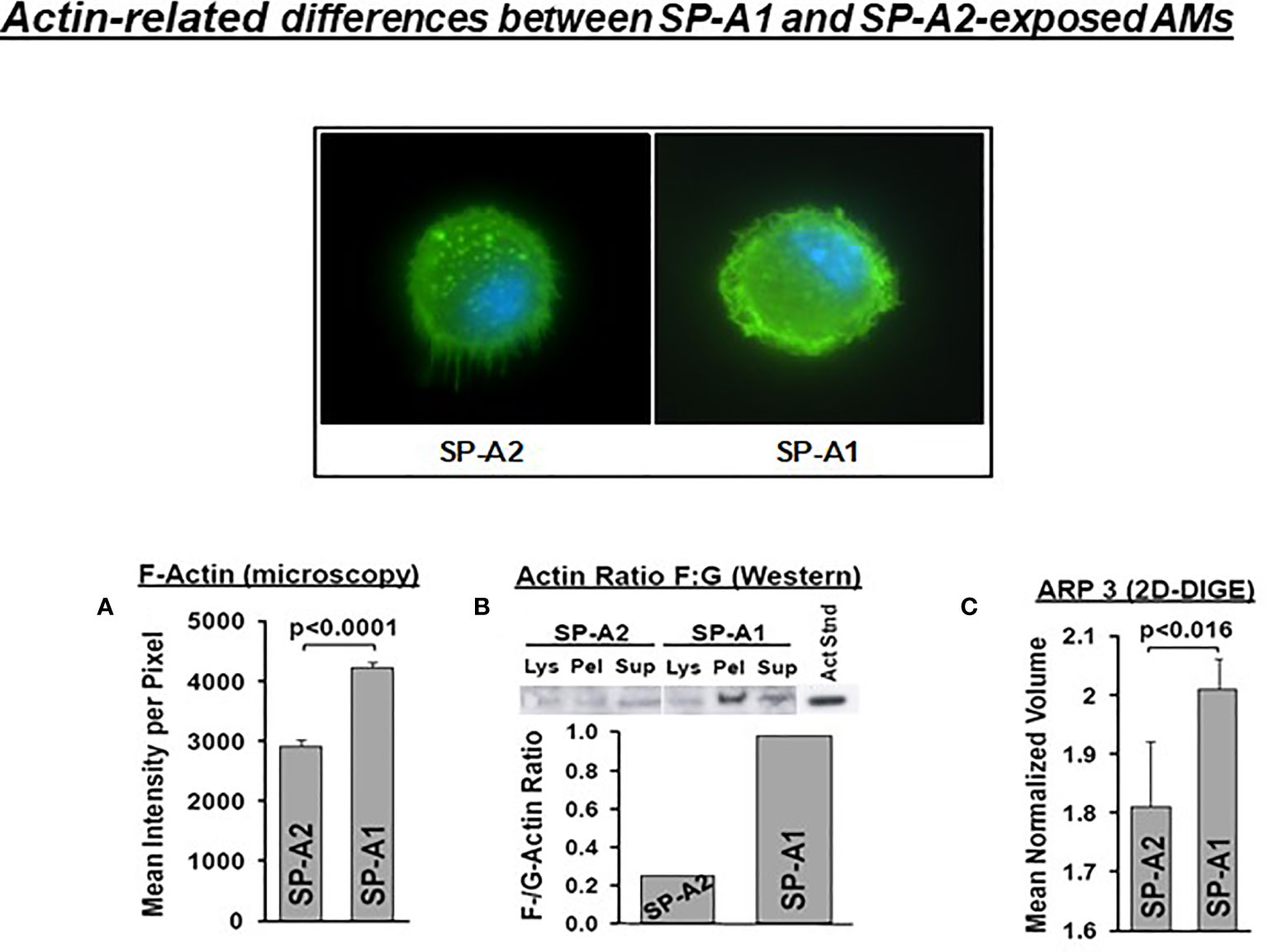
Figure 9 Actin-related differences between SP-A1 and SP-A2-exposed AM (110). The pattern of fluorescence (cells stained with AlexaFluor 488 phalloidin) of AM exposed chronically to SP-A2 or SP-A1 is shown and it is not identical. (A) depicts the mean intensity of fluorescence per pixel and shows significant differences between SP-A2 and SP-A1 AM. (B) shows the ratio (densitometric measurements) of F-actin to G-actin in different cell fractions from a Western blot stained with an anti-actin N-terminal antibody (Sigma #2103). This ratio is lower in SP-A2 AM. (C) depicts significant differences between SP-A1 and SP-A2 in the levels of ARP3 obtained from proteomics data. Arp3 regulates actin polymerization and is part of the Arp2/3 complex, where several signaling pathways leading to actin polymerization converge.
Another study employing an imaging approach of single cell analysis using fluorescence and confocal microscopy showed AM population heterogeneity (116). The F-actin cytoskeleton assessment, based on the fluorescence intensity of phalloidin staining, revealed a number of AM phenotypes (A-D). These phenotypic subpopulations differed in an SP-A genotype-, sex-, and age-dependent manner (Figure 10), as well as to whether AM were exposed to SP-A1 or SP-A2 chronically or acutely (not shown). The G-actin intensity changed among the F-actin-based AM subpopulations, and measurements indicated that these cell phenotypic responses were likely due to changes in gene expression rather than simply cytoskeleton remodeling (116). In old male SP-A1 or SP-A2 mice, the AM subpopulation associated with AM inactivation was increased, but this was not quite the case in females. In young mice the frequency of each of these actin-based phenotypes was the same in SP-A2 and co-ex females indicating that the presence of SP-A1 (in co-ex) may not have any major effect on the phenotypes compared to those from AM exposed to SP-A2 only (116). Similarities as well as differences of the impact of SP-A2 vs co-ex in various read-outs have been shown with the AM miRNome (117) and survival after bacterial infection (118), as well as with proteomics where the SP-A2 AM profile was most similar to wild type (that may be considered closer to co-ex), especially for actin cytoskeleton proteins and proteins regulated by Nrf2 (112). Collectively, these indicate that SP-A2 may be the driving force for the determination of AM phenotype especially in host defense processes. While there is no direct evidence of the functional significance of these subpopulations, the speculation that they reflect different stages of activation seems reasonable, and is supported by studies addressing the role of actin in polarity of macrophage function in different systems. Inhibition of ROCK kinase, a key regulator of the actin cytoskeleton, affected macrophage polarity and function (119). Importantly, G-actin was identified as a potential target of macrophage polarization in anti-tumor therapy (119).
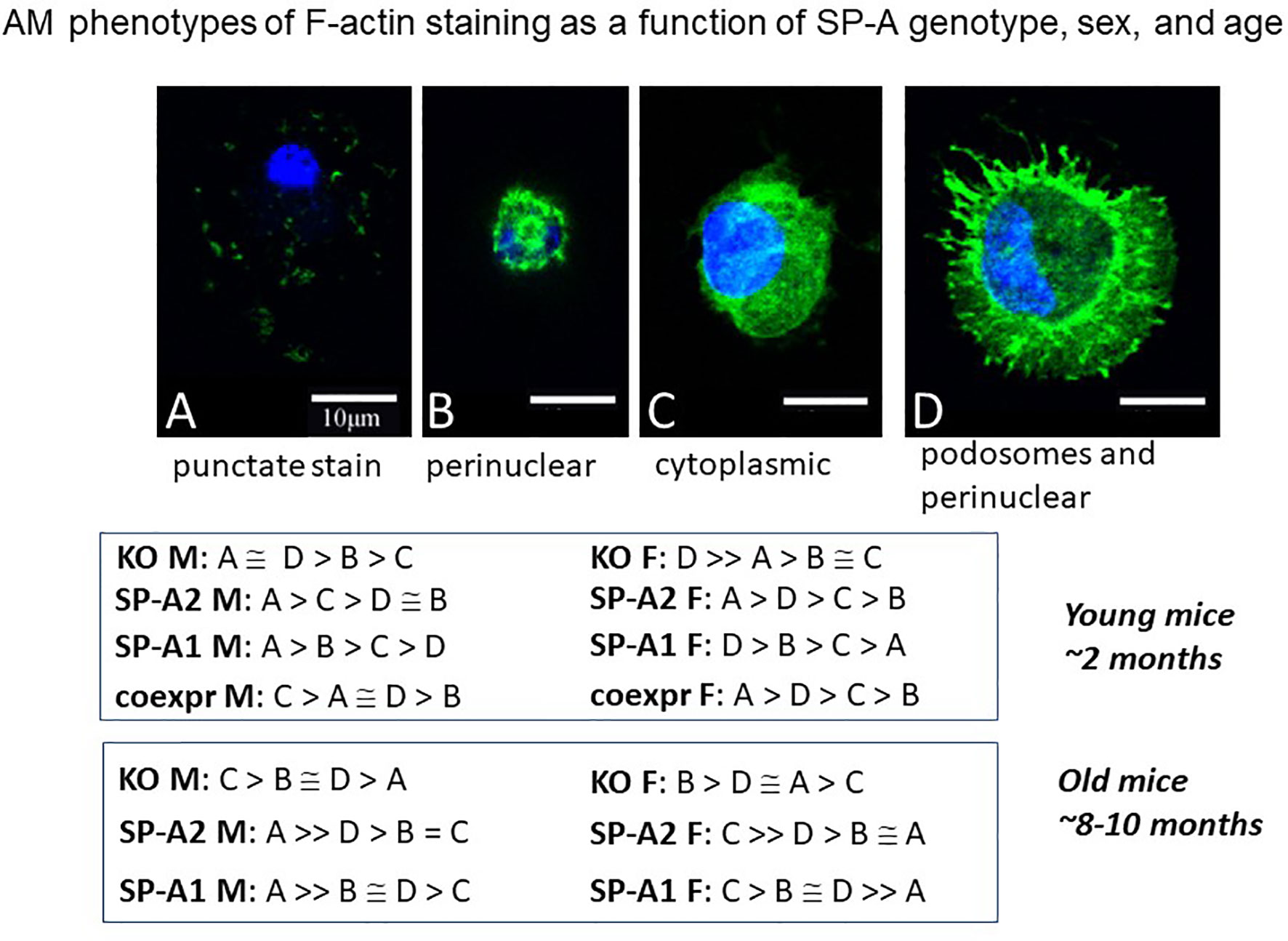
Figure 10 AM phenotypes of F-actin staining as a function of SP-A genotype, sex, and age. The fluorescence pattern of four types of F-actin staining is shown. The F-actin stain in (A) is punctate i.e. scattered cytoplasmic, in (B) perinuclear, in (C) diffuse in cytoplasm, and in (D) cortical with cytoplasmic protrusions (podosomes) and perinuclear. Cells were fixed, permeabilized and stained with Alexa Fluor 488-conjugated phalloidin. Images were obtained with a Leica SP8 AOBS laser scanning confocal system with software-adjusted detection spectra to avoid bleed through of the signal as described in detail by Tsotakos et al. (116). The relative frequency of these types in young and old mice are shown as a function of SP-A genotype and sex.
Actin cytoskeleton dynamics are critical for the main functions of AMs, such as phagocytosis (78, 79, 85), and phenotypic differences may reflect differences in the activation status of the AMs (116). While this assumption has yet to be confirmed, a novel subpopulation of AMs (CD11c+ CD11b+) was described in the BAL of old mice. These AMs display pro-inflammatory characteristics and are permissive to Mycobacterium tuberculosis (120). The lung environment may control the metabolic status of these cells, thereby affecting their activation status (121). Currently it is not known whether and how SP-A1 and SP-A2 may differentially affect AM responsiveness to M. tuberculosis, but numerous studies (as discussed in this review) have shown that the two gene products differentially affect nearly all the AM read-outs studied to date, with SP-A2 being more effective in the enhancement of innate immune-related responses, such as pro-inflammatory responses, as well as morphological changes, as seen by the actin-related proteome differences (114).
6b. Alveolar Cell miRNome
SP-A1 (6A2) and SP-A2 (1A0) differentially regulate the miRNome of the epithelial Type II cell (122) and the AM (123) in a genotype-dependent and sex-specific manner at 4hr post O3 exposure and after normalization to the corresponding SP-A KO groups. SP-A2 had a significant impact on the AM miRNome and SP-A1 on the epithelial Type II miRNome. In both cell types the miRNome of males (but not females) changed significantly in response to O3 exposure. Gonadectomy had a major impact on the miRNome of males when compared to females under both control conditions (filtered air-exposed mice) and after O3 exposure, indicating a role of sex hormones. A considerable amount of evidence exists that sex hormones do indeed control immune response (124).
The SP-A2-mediated changes in the AM miRNome in males showed (at 4hr post O3 exposure) their miRNA target genes to be involved in pro-inflammatory, anti-apoptotic, and antioxidant pathways (123). But when both SP-A1 and SP-A2 gene products were expressed (co-ex) in the same humanized transgenic (hTG) mouse, both similarities and differences were observed in the specific AM miRNA-targeted genes and pathways when compared to hTG SP-A2 male mice under similar conditions (i.e., at 4hr post O3 exposure) (117). The common pathways included the pro-inflammatory response (STAT3, NF-kB) and the anti-apoptosis pathway (117). However, in males expressing only SP-A2, the reactive oxidant species (ROS) homeostasis pathway was involved, whereas in males expressing both SP-A1 and SP-A2 proteins the cell cycle, growth, and proliferation pathway was involved (123). A recent study where the NAD(H) redox status of AM after O3 exposure was investigated using optical redox imaging showed SP-A2 to be involved in processes of ROS homeostasis (103). AMs from males were more oxidized and had higher mitochondrial ROS than females. Together these indicate that AM from SP-A2 males are more sensitive/responsive to O3 exposure than females, and as a result there may be a more robust regulation of the AM male miRNome and its miRNA-targeted genes involved in ROS-homeostasis pathway. But when both SP-A1 and SP-A2 proteins are present perhaps a more sustained recovery is underway, as assessed by the finding where the regulated AM miRNAs and their target genes are involved in cell cycle, growth, and proliferation processes. In co-ex, although a downregulation was largely observed in the miRNome of both male and female mice, the expression of target genes studied was opposite in males (upregulation) and females (downregulation) (117). The SP-A2 males showed overall similarities with the co-ex males where there was largely a downregulation of the miRNome and upregulation of the target genes (117, 123). Moreover, differences in the AM miRNome and their target genes between co-ex and SP-A2 male and female mice persists even at 18hr post O3 exposure, with O3 exposure shown to attenuate sex differences (125). At this time point the overall pathways were similar but the direction and the number of changed miRNAs in response to O3 varied, indicating underlying complexities in the mechanisms involved and an interplay of O3 exposure, time of post exposure, sex, and SP-A genotype.
6c. AM Gene Expression
Differences between SP-A1 (6A2, 6A4) or SP-A2 (1A0, 1A3) variants, and KO in response to infection are observed in the regulation of AM gene expression in males and females (126). Ingenuity Pathway Analysis (IPA) revealed key pathways and molecules involved in TP53, TNF, and cell cycle signaling nodes. All variants, except SP-A2 (1A3) female, showed significance for at least two of the three pathways studied, and the KO male showed significance for all three pathways. The expression profile of validated genes was sex- and variant-specific, and a similarity in the gene expression profile of KO and SP-A1 mice was observed (126).
6d. Toponomics of Alveolar Macrophages
The presence of SP-A affects multiple functions or cellular states of the AM, as discussed above. In general, in nearly all quantitative studies, a number of cells are combined or pieces of tissue are used, so differences between individual cells are lost, and the specific read-out usually either increases or decreases or remains unchanged. However, this by itself cannot provide any insight regarding “patterns of protein combinations” in a macromolecular cluster within individual cells. The Toponome Imaging System (TIS), a serial immunostaining system combining aspects of the proteome and interactome, permits the co-localization of multiple proteins within intact single cells, and allows the identification and enumeration of supramolecular structures formed by protein clusters, known as combinatorial molecular phenotypes or CMPs (127–131). CMPs are likely the basis of cellular function because proteins usually function as part of a complex rather than alone. An example of a pseudo-colored image of cells with colors corresponding to each CMP within a given single AM is shown in Figure 11. This provides a visual depiction of the diversity of AM, where no two AMs are identical. AMs from SP-A1, SP-A2, or KO mice exhibit an extensive CMP-based diversity (132) that may translate into functional differences and serve as the basis of the SP-A variant-dependent effects in AMs described above, and shown previously in other systems (127, 133, 134). We found SP-A1 AMs to be more heterogeneous than either KO or SP-A2 AMs. A similar CMP-based AM heterogeneity was observed previously in AM from SP-A KO and SP-A1-rescued KO mice (135), where a single SP-A1 treatment of SP-A-KO mice increased CMP-based diversity compared to those treated with vehicle. The CMPs in KO were more conserved (i.e., present in all or most members of the experimental group) than in SP-A1 treated cells. The less abundant CMPs were more diverse i.e., these were composed of a greater number of markers. The CMP data analysis clearly showed that “patterns of protein combinations” between the two groups (presence or absence of SP-A1) are not all-or-nothing and certain CMPs are predominant in one or the other group and some are unique to one or the other group. This rheostat-like CMP presence may have functional consequences and be the basis of SP-A1- and SP-A2-mediated functional and regulatory differences observed in AMs discussed above. Our unpublished studies also show sex-dependent CMP differences in AM. Combined, these show a role for SP-A in AM diversity based on CMPs. As the proteins in a given CMP hold the potential to interact with one another to mediate or contribute to a given function, CMP differences may underlie not only the SP-A variant-mediated AM function and regulation discussed above, but other roles yet to be described.
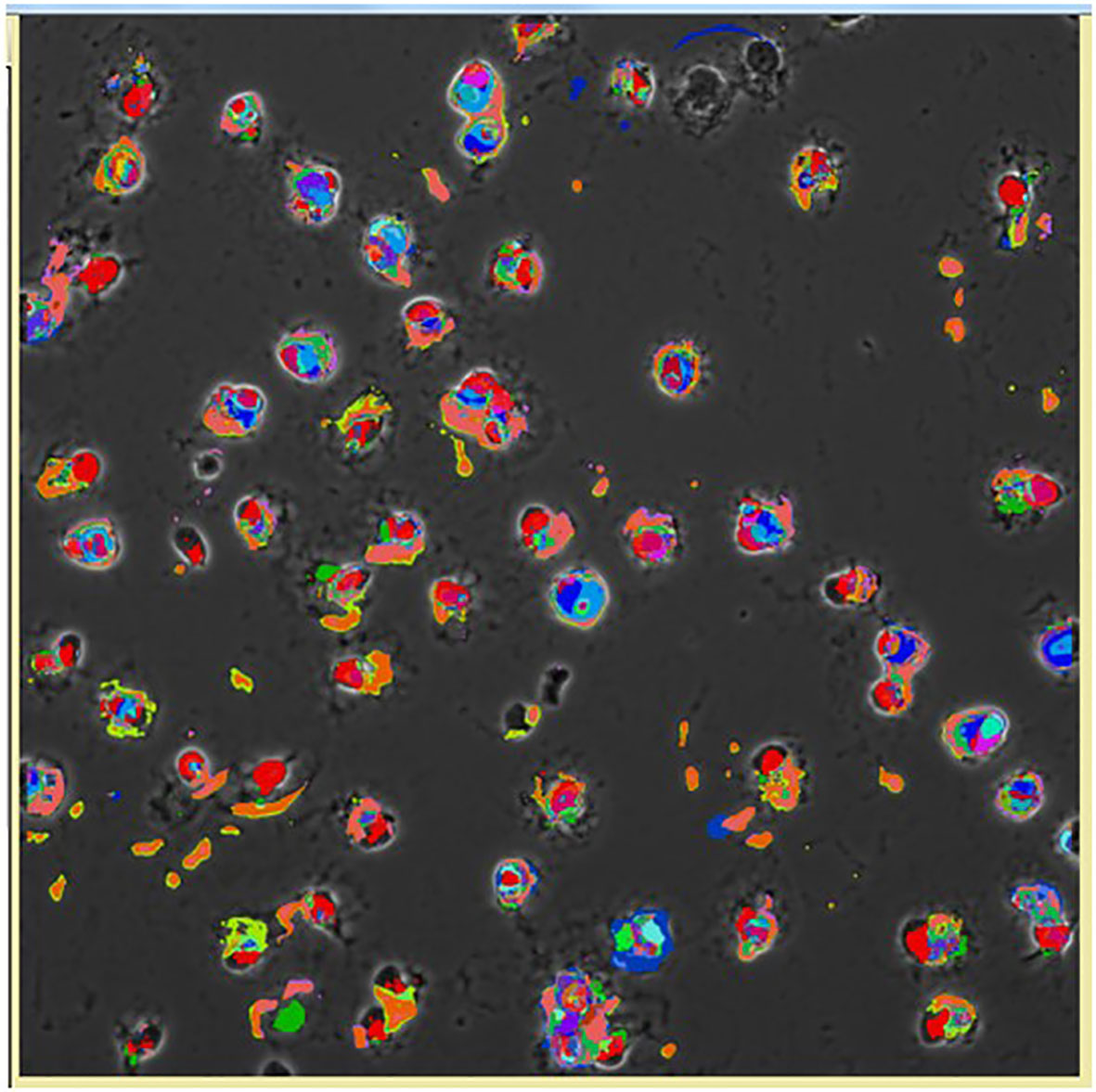
Figure 11 Pseudocolored image of cells based on the CMPs within each cell using the Toponome imaging system. Pseudocolors correspond to each CMP present in the given cell and are assigned by the Multicompare program. These pseudocolors are then superimposed on the appropriate phase contrast image as described in detail by Phelps et al. (132). Please note that no two cells are identical.
7. Functional Read-Outs in the Intact Organism
7a. Lung Function and Survival in SP-A1 and SP-A2 hTG Mice
hTG mice, each expressing a different SP-A1 or SP-A2 variant, revealed differences in airway lung function read-outs after K. pneumoniae infection. In the absence of methacholine, sex differences were observed for most parameters studied, with no significant differences being observed between SP-A1 and SP-A2 variants. In the presence of methacholine both sex- and variant-dependent differences were observed (136). The SP-A2 (1A3) exhibited increase in the lung function read-outs in both sexes when compared to SP-A2 (1A0), and the SP-A1 (6A2, 6A4) showed more diverse lung functional responses.
Survival following K. pneumoniae infection differed among young (~12 weeks old) hTG mice expressing different SP-A gene variants. Mice expressing one or both (co-ex) SP-A transgenes had significantly increased survival rates following infection compared to KO (co-ex=SP-A2 > SP-A1 > KO) (118). Survival rate differences among SP-A1 and SP-A2 variants were observed with co-ex (6A2/1A0) and SP-A2 (1A0) exhibiting the best survival: co-ex (SP-A1 (6A2)/SP-A2 (1A0)) = SP-A2 (1A0) > SP-A2 (1A3) = SP-A1 (6A2) > SP-A1 (6A4) > KO (118). This order is reminiscent of the one obtained in the ex vivo phagocytosis assays (85) described above. In all cases (except for co-ex where there was no significant sex difference) females exhibited a better survival than males. For variants 6A4, 6A2, and 1A3, males had a significantly poorer survival compared to co-ex, females (6A2, 1A3), or 1A0 (males or females) (118). The lack of sex difference in co-ex indicates that expression of both gene products at similar levels, as it is in this experimental model, may eliminate the male survival disadvantage following bacterial infection. However, this may not be the case if the relative levels in BAL of SP-A1 and SP-A2 vary significantly. Variable levels in BAL were seen in the ratio of SP-A1 to total SP-A in healthy subjects and in various patients; the ratio of SP-A1 to total SP-A was increased in subjects with asthma (137), and in cystic fibrosis and bacteria positive samples (73). Although this ratio is not based on absolute levels due to the different affinities of the antibodies used, it is however a useful tool to indicate that differences do exist in health and disease and perhaps also as a function of age (73). The details of the SP-A1 antibody generation and specificity as well as the assays used are described by Tagaram et al. (73). Following these studies an SP-A2-specific antibody became commercially available (126). The SP-A antibody that recognizes both SP-A1 and SP-A2 was generated (87) against purified SP-A (138) from alveolar proteinosis. Future studies are needed using antibodies with equal affinities, to determine the range of absolute values in health and disease and/or among various populations.
In aged mice (~9-12 months) a significantly better survival after K. pneumoniae infection was observed in mice expressing the SP-A2 (1A0) variant with no major variant- or sex-specific differences being observed among the other variants, except for the SP-A1 (6A2) that showed better survival in females compared to males (139). A serendipitous and perhaps a clinically relevant observation (140) was made in the course of this survival study. Improved ventilation, as provided by the experimental conditions (i.e., high flow of humidified, warmed (37.5°C) and at constant pressure of 1.5 cm H2O filtered air), resulted in a significantly better survival of aged infected mice compared to those with infection alone and without prior improved ventilation; The SP-A2 (1A0) provided a better overall survival in both males and females. The improved survival with prior ventilation held true for young mice (~12 weeks old), but the magnitude of change was considerably smaller in young mice compared to aged mice (~9-12 months old) (139). O3 exposure prior to infection had an overall negative impact on the survival of aged mice compared to those with improved ventilation prior to infection. A gene-, sex-, and variant-specific survival was observed in the O3-exposed group that was subjected to improved ventilation prior to infection. The SP-A2 variants were associated with better survival in males and the SP-A2 (1A0) variant with better survival in females compared to the SP-A2 (1A3) variant.
The SP-A variant-, exposure-, and age-dependent survival clearly indicates that SP-A has a significant positive effect. Although a single SP-A dose led to a significantly better survival after infection, it is not known which SP-A variant (if used for therapy) could positively affect survival in the presence of additional environmental insults. Thus, it would be of interest to identify SP-A variants or functional SP-A fragments that, for example, could resist O3-induced oxidation and thus maintain all or most of their functional capacity under those conditions.
7b. Survival in Humans After Lung Transplantation as a Function of SP-A Genetics
In humans, after lung transplantation, survival was significantly better in the first year, which is the most critical time for these patients, if the SP-A2 genotype of the donor lung was 1A0:1A0 compared to the SP-A2 1A0:1A1 genotype. The SP-A2 (1A0), versus other SP-A2 variants, was also associated with better survival in post-transplant patients (141), as predicted by the survival of young (118) and aged (139) hTG mice carrying different SP-A1 and SP-A2 variants. Moreover, precision-cut lung slices from human donor lung following incubation with methylprednisolone, showed that lungs carrying the SP-A2 (1A0) variant had higher levels of SP-A compared to lungs carrying other SP-A variants (142). Low SP-A levels have been shown to associate with poor outcome and early lung transplant survival (143). Collectively, the available data indicate an important role of SP-A genetics for optimal lung health and survival in lung transplant patients.
8. Association of Human Surfactant Protein A Variants With Pulmonary Disease
The importance of the human SP-A genetics in human disease is underscored by the large number of association studies, where SP-A1 and SP-A2 variants or single SP-A1 and SP-A2 nucleotide polymorphisms have been shown individually or via interactions between or among themselves to associate with disease susceptibility (risk or protection) or disease severity. These studies have been reviewed and described in detail elsewhere (144–147). However since the publication of these review articles, the list of such studies continues to grow (95, 148–153). This is not surprising because innate host defense, regulation of inflammatory processes and/or surfactant dysregulation/derangement are central to most (if not to all) pulmonary diseases and SP-A is shown to play a role in these processes. Thus, a better understanding of the functional, biochemical, regulatory and other differences among the SP-A genetic variants in health and disease is of the utmost importance if we were to improve disease-specific and patient-specific decision making in the clinic and consider potential SP-A-specific therapies for various patient groups or individuals. The significantly improved survival of bacterial infected SP-A KO animals after a single dose of SP-A treatment (118) indicates the feasibility of such studies and as discussed in the last section of this review, efforts are underway in preclinical studies towards the use of SP-A therapeutically. Moreover, the potential role of these variants in infectious diseases such as in COVID-19 has been discussed elsewhere (154, 155).
9. Discussion
If the gold standard for an innate immune molecule were to be survival of the organism in response to an insult (i.e., infection), then SP-A is an important host defense molecule in the lung. The survival of young SP-A KO mice after infection and/or certain processes associated with in vivo infection are considerably poorer compared to wild type animals (100, 156–159). Moreover, a single dose of exogenous SP-A treatment of SP-A KO mice prior to infection, after infection, or at the time of infection significantly improved survival (118). The overall survival after infection, regardless of SP-A variant, in most cases depending on the sex of the animal, is significantly higher than the KO (118), even though no significant difference is observed among the various SP-A mouse lines without infection. A cartoon depicting this SP-A variant- and sex-dependent survival outcome is shown in Figure 12. These observations indicate, not only that SP-A is key for effective lung innate immunity, but that the SP-A genetics variably alter outcome and thus are likely to contribute to lung disease susceptibility.
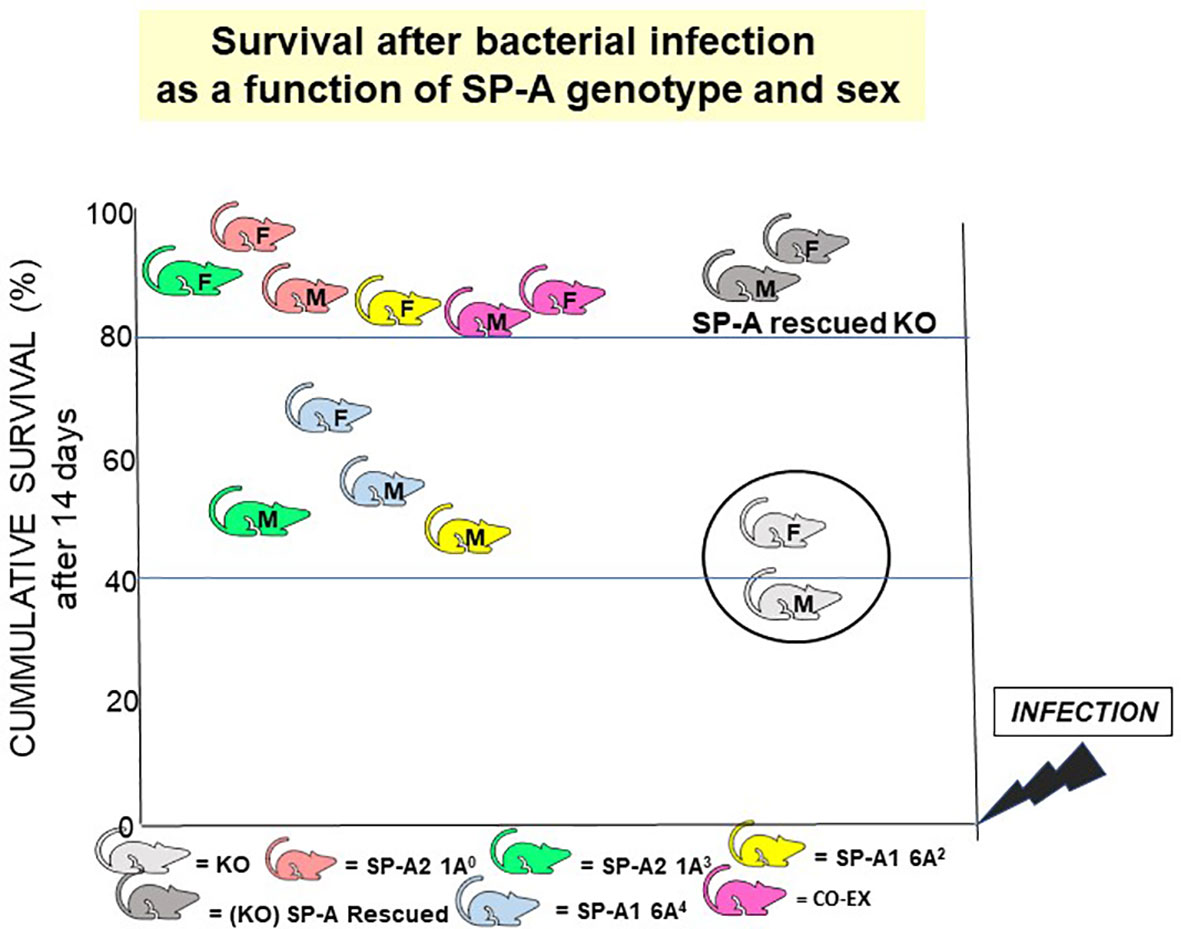
Figure 12 SP-A genotype- and sex-dependent survival after bacterial infection. This cartoon shows that: a) in the absence of SP-A i.e., KO, survival after Klebsiella pneumoniae infection is poor but survival is significantly improved if the KO mice are rescued with SP-A; and b) survival differs as a function of SP-A genotype and sex (118). However, in the absence of infection, no significant differences have been observed in mice expressing different variants or in SP-A KO mice, if the mice are housed in a pathogen-free environment.
Age and O3 exposure prior to infection have a negative impact on survival (100, 139). Observed differences in survival among variants of infected young mice (118), get minimized or eliminated in aged infected mice after O3 exposure (139). Of interest, the higher survival observed with the SP-A2 (1A0) variant in aged mice, was also observed in human lung transplant patients who tend to be of older age (149). However, a serendipitous observation (139) revealed that high flow of humidified and warmed air significantly: a) improved survival in aged mice (and young mice but to a lesser degree); and b) in certain cases this improvement was SP-A variant- and sex-dependent. Moreover, numerous studies have revealed associations between SP-A genetics variants and lung disease susceptibility (145–148, 150, 151, 160–163) further underscoring the contribution of SP-A genetics to disease susceptibility. Together these clearly indicate a differential and a beneficial role of SP-A variants. Although the observed variant-specific differences under certain conditions may be partially obliterated, the addition of other non-invasive clinical procedures, such as high flow of humidified and warmed air, improves SP-A variant- and sex-dependent survival. Whether this is the case in humans remains to be determined (140).
The question now is what happens between infection and the ultimate differential survival of mice expressing different SP-A1 and SP-A2 variants. As reviewed here several processes and functions are differentially affected by these variants in the epithelial type II cells (no studies have been done with the type I cells), the alveolar macrophage, and the hypophase i.e., the microenvironment these cells are exposed to. A cartoon is shown in Figure 13 depicting processes and/or functions of the alveolar type II cell, the hypophase, surfactant structures and the AM that SP-A1 and SP-A2 may differentially affect (Table 4). Thus, in the alveolar space, the AM, for example, as a resident cell is exposed to a different local microenvironment depending in part on SP-A genotype. At the same time the AM, and perhaps other alveolar cells, differentially contribute to the local microenvironment as a function of the SP-A variant exposure and sex. This may affect both the AM’s baseline properties and subsequently its differential response to various insults, as this may be reflected by changes in its proteome, toponome, miRNome, gene expression, or other. Rescue experiments with SP-A of SP-A KO mice support SP-A-mediated changes both at baseline conditions and in response to insults. For the former, a single dose of SP-A treatment of SP-A KO mice resulted in an AM proteome that resembled that of the WT AM and for the latter, a single dose of SP-A treatment of KO mice resulted in a better survival after infection. Moreover, SP-A genetics, as discussed, has a major regulatory impact on processes and functions of AM underscoring the importance and the complexity of the AM/SP-A interaction in health and disease.
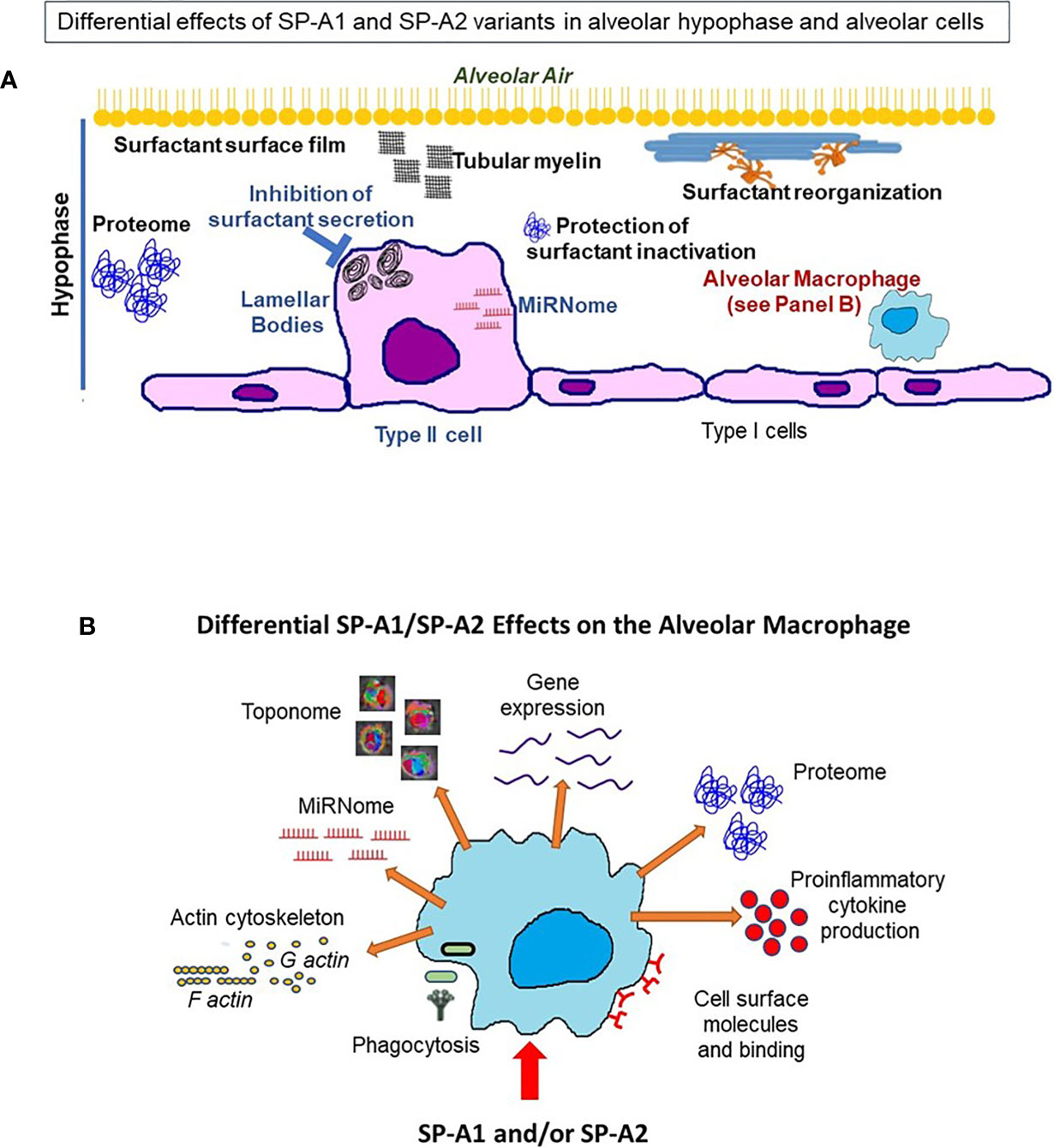
Figure 13 This cartoon provides an overview of the processes reviewed here to be differentially affected by SP-A1 and SP-A2 gene products in the alveolus, the BAL/hypophase and the alveolar cells (macrophages and epithelial cells). These processes are detailed in Table 4. (A) shows a cartoon of such processes of the type II cell miRNome, inhibition of surfactant secretion and perhaps the size of lamellar bodies. No studies involving SP-A1 and SP-A2 have been carried out for the type I cells. This panel also depicts events occurring in the hypophase. These include surfactant reorganization, surfactant surface film, proteome, and protection of surfactant inhibition by serum proteins. Both SP-A1 and SP-A2 are needed for tubular myelin formation. (B) depicts a cartoon of SP-A1 and SP-A2-mediated processes that affect AM at baseline and/or in response to insults. These include differences in AM proteome, toponome, gene expression, miRNome, bacterial phagocytosis, pro-inflammatory cytokine production, expression of cell surface molecules, AM binding and actin cytoskeleton.
In some cases, especially in the absence of SP-A, some unexpected and rather paradoxical findings were observed. These unexpected and paradoxical findings provided further insight into the various roles of SP-A in the alveolar space/cells, especially as one thinks of the SP-A’s “gold standard” outcome (i.e., survival of the organism after infection). Such examples include the following. The BAL proteome from SP-A KO animals exposed to filtered air was reminiscent of that from wild type animals in response to O3 exposure (102) indicating that in the absence of SP-A, the KO may be in a state of chronic oxidative stress. This now has been confirmed in a recent study where the redox status of KO and SP-A2 AM was assessed using a redox imaging technique and was shown to be sex-dependent (103). Also, in the same study SP-A2 in a sex-dependent manner affected the NAD(H) redox homeostasis of the AM in response to both filtered air and O3 exposure (males) or O3 exposure (females). Another interesting observation is that following analysis of the KO and WT BAL proteome after bacterial infection. Certain proteins involved in host defense were upregulated in SP-A KO BAL under baseline conditions, indicating perhaps, an attempt to overcome host defense deficits in the absence of SP-A. However, these were decreased after infection and the expression of other proteins involved in host defense were altered in KO compared to WT. These changes, because they do not, as shown by survival studies of infected mice, translate into survival rates seen in WT mice, indicate a dysregulation in one or more SP-A-mediated processes discussed in this review.
Whether the two SP-A gene products and their corresponding variants are equivalent in function, structure, or other (164), after evaluation of the collective literature, it is clear that the SP-A1 and SP-A2 gene products exhibit both unique and overlapping function/activity with extensive qualitative differences among variants. Each SP-A genotype exhibited differences from one another in nearly all the read-outs studied. Table 4 summarizes a number of functions where one SP-A gene product exhibits a more robust response, higher functional activity, or various characteristics that differ from the other. However, SP-A2 shared more similarities with the co-ex (presence of both SP-A1 and SP-A2) in a sex-specific (or not) manner, in several of the read-outs, especially, the ones associated with or holding the potential to regulate innate immune responses. These may include studies relating to survival, AM miRNome, proteome, actin cytoskeleton, and other. In some cases, the pattern of expression or regulation as, for example, was shown for the AM proteome, actin-related pathways, and others, was opposite between SP-A1 and SP-A2. The SP-A1, on the other hand, compared to other SP-A genotypes (SP-A2, KO) exhibited more diverse patterns (i.e., less consistency/similarity). For example, in the AM gene expression pathways, SP-A1 exhibited some similarity with the KO AM, whereas with regards to the AM CMP-based diversity, SP-A1 showed more heterogeneity than either the KO or SP-A2. SP-A1 also had an inhibitory effect on AM binding by SP-A2, and a lower ability to self-aggregate by itself or in the presence of SP-A2, but a better efficiency in surfactant lipid membrane reorganization, plus a better protection against surfactant inactivation by serum proteins.
In summary, SP-A variants differentially affect the BAL proteome, and consequently the local microenvironment where the sentinel lung innate immune cell, AM, resides, and with which SP-A interacts to regulate several of its processes and functions (Figure 13B and Table 4). In other words, SP-A variants may not only differentially affect AM processes and function in response to various insults, but also the status of AM “readiness” to respond to various insults. In fact, analysis of combinational molecular phenotypes (CMPs) in individual AM cells under unprovoked conditions showed that AM exposed continually in vivo to different SP-A variants exhibit an extensive and variable CMP-based diversity. This CMP diversity may be the basis of SP-A variant-mediated effects observed in AM. As single CMPs or groups of CMPs may be poised to mediate a specific function in the cell, we postulate that some of the unexpected findings in SP-A KO are partly the result of the shared CMPs observed between SP-A KO and AM exposed to SP-A1 or SP-A2. However, the ultimate positive functional outcome or maximal survival can only be realized when SP-A is present to mediate the formation of specific groups of CMPs necessary for a better outcome. Moreover, SP-A variants individually may differentially affect this CMP complexity resulting in the overall SP-A gene- and variant-specific survival differences observed. Furthermore, SP-A variants have been shown to differentially regulate the miRNome of alveolar type II epithelial cells (122) and inhibition of ATP-stimulated phosphatidylcholine secretion by Type II cells (23) (Figure 13A). However, no other studies have been done where the impact of SP-A genetics on various aspects of the alveolar epithelial Type II or Type I cells was investigated. Thus, additional mechanisms may mediate changes in the local microenvironment that may control the metabolic status of the alveolar cells, thereby affecting their functional/activation status (121). These may involve, either SP-A variant-mediated processes not yet studied or non-SP-A mediated processes, or both.
On a final note, the use of SP-A in the clinic, and especially in the prematurely born infants who may develop respiratory distress syndrome or bronchopulmonary dysplasia has not been considered (165). These infants exhibit not only pulmonary surfactant deficiency (166), but also SP-A deficiency (67, 167, 168). Although surfactant replacement therapy is routinely used, none of the preparations contains SP-A even though a major complication in these infants is infection. Based on the available literature, inclusion of SP-A, or biologically functional fragments of SP-A in the existing surfactant preparations or SP-A usage alone must be considered for treatment of this patient population. Prematurely born infants historically constitute the first patient population where surfactant deficiency was recognized as a problem (169) and where the study of surfactant proteins began. Currently, surfactant SP-A therapy could be used, beyond the prematurely born infant, in several other diseases of infectious origin or diseases caused by exposure to various irritants/allergens. The fact that a single dose of SP-A has a major impact on animal survival and the AM proteome speak to the feasibility of SP-A therapy use in the clinic. Moreover, the potential role of the SP-A variants in infectious diseases such as COVID-19 has been recently discussed (154, 155). Furthermore, as reviewed here, SP-A may have a positive impact beyond innate immunity by benefitting surfactant-related functions, not only in prematurely born infants with neonatal disease, but in other pulmonary diseases. Of interest, trimeric fragments of human SP-A have been used successfully to neutralize RSV infection in an in vitro model of human bronchial epithelial cells (66). Furthermore, in animal models and human epithelial cells, exogenous SP-A is shown to attenuate asthma-related factors and protect against IL-13-induced inflammation in asthma (170).
10. Conclusions/Summary
The SP-A1 and SP-A2 variants, in an SP-A genotype-, sex-, age-, and condition-specific manner, differentially affect multiple read-outs in: a) the AM that may include its proteome, miRNome, gene expression, toponome, bacterial phagocytic index, cytoskeleton, and ability to produce pro-inflammatory cytokines; b) bronchoalveolar lavage (proteome); c) epithelial Type II cells (miRNome); d) surfactant structure and re-organization, and inhibition of surfactant inactivation by serum proteins; e) parameters of lung mechanics; f) survival of young and aged mice after bacterial infection with or without O3 exposure; and g) survival of lung transplant patients. In addition, the SP-A1 and SP-A2 variants differ in their ability to form different size oligomers and self-aggregate as well as in their sensitivity to oxidation. Thus, these differences collectively indicate that the genetics of the innate immune molecules, SP-A1 and SP-A2, and their corresponding variants, may play an important role in lung health under various conditions and this postulate is supported by the large number of studies where associations between SP-A variants and various pulmonary diseases have been observed. Furthermore, the collective evidence indicates that SP-A2 is more effective in the enhancement of innate immune-related responses and shares more similarities in several read-outs with the co-ex, where both SP-A1 and SP-A2 are expressed in the same mouse. On the other hand, SP-A1 exhibits higher efficiency in surfactant structure re-organization and in the inhibition of surfactant function by serum proteins, whereas both proteins appear to be necessary for the formation of the extracellular structural form of surfactant, tubular myelin. While the mechanisms leading to the differential effect of SP-A1 and SP-A2 on the function of AMs have not been elucidated, the initial steps may include the differential ability of SP-A1 and SP-A2 to bind to receptors in the surface of AMs. SP-A2 exhibits a higher ability to bind phagocytic cells, such as AMs and THP-1 cells (a phagocytic macrophage-like cell line) than SP-A1, but no differences were observed between SP-A1 and SP-A2 in the binding of CHO cells, a non-phagocytic cell line (118).
Author Contributions
JF wrote most of the review, integrated all parts and figures and wrote the discussion. NTh wrote parts of the review and the Tables. NTs wrote parts of the review and contributed to some of the figures. DP wrote parts of the review and contributed to some of the figures. All authors contributed to the article and approved the submitted version.
Funding
Supported by the CHILD Fund and the John Ardell Pursley Memorial Research Fund, Department of Pediatrics, Penn State University College of Medicine.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Dr. Jesus Perez-Gil and his lab for making available published figures and Dr. Shaili Amatya for her contributions towards Figures 12 and 13.
Abbreviations
AM, Alveolar macrophage; ARP, acute phase response; BAL, Bronchoalveolar lavage; CCT2, chaperonin subunit (beta); CMP, combinational molecular phenotypes; CRD, C-terminal carbohydrate recognition domain; DPPC, dipalmitoyl phosphatidylcholine; DPPG, dipalmitoyl phosphatidylglycerol; hTG, humanized transgenic mice; KO, Knockout; K. pneumoniae, Klebsiella pneumoniae; LPS, lipopolysaccharide; MBL3P, mannose-binding lectin family member 3 pseudogene; miRNA, micro RNA; M. tuberculosis, Mycobacterium tuberculosis; NAD(H), Nicotinamide adenine dinucleotide; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B cells; Nrf2, Nuclear factor erythroid 2-related factor 2; O3, ozone; ROS, reactive oxygen species; SFTPA1, Pulmonary surfactant-associated protein A1 gene; SFTPA2, Pulmonary surfactant-associated protein A2 gene; SP-A, Surfactant Protein A; SP-A1, Surfactant Protein A1; SP-A2, Surfactant Protein A2; SP-B, Surfactant Protein B; SP-C, Surfactant Protein C; SP-D, Surfactant Protein D; STAT-3, Signal transducer and activator of transcription 3; STPA3P, surfactant protein A3 pseudogene; TIS, Toponome Imaging System; WT, wild type.
References
1. Autilio C, Perez-Gil J. Understanding the Principle Biophysics Concepts of Pulmonary Surfactant in Health and Disease. Arch Dis Childhood Fetal Neonatal edition (2019) 104:F443–51. doi: 10.1136/archdischild-2018-315413
2. Floros J, Phelps DS. editors: Yaksh TL, Lynch C III, Zapol WM, Maze M, Biebuyck JF, Saidman LJ. Lippincott-Raven Publishers. Philadelphia: Anesthesia: Biologic Foundations (1997). p. 1259–80.
3. Hoover RR, Floros J. Organization of the Human SP-A and SP-D Loci at 10q22-Q23. Physical and Radiation Hybrid Mapping Reveal Gene Order and Orientation. Am J Respir Cell Mol Biol (1998) 18:353–62. doi: 10.1165/ajrcmb.18.3.3035
4. Sastry K, Herman GA, Day L, Deignan E, Bruns G, Morton CC, et al. The Human Mannose-Binding Protein Gene. Exon Structure Reveals Its Evolutionary Relationship to a Human Pulmonary Surfactant Gene and Localization to Chromosome 10. J Exp Med (1989) 170:1175–89. doi: 10.1084/jem.170.4.1175
5. Floros J, Hoover RR. Genetics of the Hydrophilic Surfactant Proteins A and D. Biochim Biophys Acta (1998) 1408:312–22. doi: 10.1016/S0925-4439(98)00077-5
6. Cañadas O, Olmeda B, Alonso A, Pérez-Gil J. Lipid-Protein and Protein-Protein Interactions in the Pulmonary Surfactant System and Their Role in Lung Homeostasis. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21103708
7. Wright JR. The “Wisdom” of Lung Surfactant: Balancing Host Defense and Surface Tension-Reducing Functions. Am J Physiol Lung Cell Mol Physiol (2006) 291:L847–50. doi: 10.1152/ajplung.00261.2006
8. Possmayer F, Nag K, Rodriguez K, Qanbar R, Schurch S. Surface Activity In Vitro: Role of Surfactant Proteins. Comp Biochem Physiol Mol Integr Physiol (2001) 129:209–20. doi: 10.1016/S1095-6433(01)00317-8
9. Schurch S, Green FH, Bachofen H. Formation and Structure of Surface Films: Captive Bubble Surfactometry. Biochim Biophys Acta (1998) 1408:180–202. doi: 10.1016/S0925-4439(98)00067-2
10. Perez-Gil J, Keough KM. Interfacial Properties of Surfactant Proteins. Biochim Biophys Acta (1998) 1408:203–17. doi: 10.1016/S0925-4439(98)00068-4
11. Echaide M, Autilio C, Arroyo R, Perez-Gil J. Restoring Pulmonary Surfactant Membranes and Films at the Respiratory Surface. Biochim Biophys Acta Biomembranes (2017) 1859:1725–39. doi: 10.1016/j.bbamem.2017.03.015
12. Palaniyar N, Ikegami M, Korfhagen T, Whitsett J, McCormack FX. Domains of Surfactant Protein A That Affect Protein Oligomerization, Lipid Structure and Surface Tension. Comp Biochem Physiol Mol Integr Physiol (2001) 129:109–27. doi: 10.1016/S1095-6433(01)00309-9
13. Zuo YY, Veldhuizen RA, Neumann AW, Petersen NO, Possmayer F. Current Perspectives in Pulmonary Surfactant–Inhibition, Enhancement and Evaluation. Biochim Biophys Acta (2008) 1778:1947–77. doi: 10.1016/j.bbamem.2008.03.021
14. Casals C, Miguel E, Perez-Gil J. Tryptophan Fluorescence Study on the Interaction of Pulmonary Surfactant Protein A With Phospholipid Vesicles. Biochem J (1993) 296(Pt 3):585–93. doi: 10.1042/bj2960585
15. Garcia-Verdugo I, Canadas O, Taneva SG, Keough KM, Casals C. Surfactant Protein A Forms Extensive Lattice-Like Structures on 1,2-Dipalmitoylphosphatidylcholine/Rough-Lipopolysaccharide-Mixed Monolayers. Biophys J (2007) 93:3529–40. doi: 10.1529/biophysj.107.109793
16. Ruano ML, Nag K, Casals C, Perez-Gil J, Keough KM. Interactions of Pulmonary Surfactant Protein A With Phospholipid Monolayers Change With pH. Biophys J (1999) 77:1469–76. doi: 10.1016/S0006-3495(99)76994-8
17. Ruano ML, Nag K, Worthman LA, Casals C, Perez-Gil J, Keough KM. Differential Partitioning of Pulmonary Surfactant Protein SP-A Into Regions of Monolayers of Dipalmitoylphosphatidylcholine and Dipalmitoylphosphatidylcholine/Dipalmitoylphosphatidylglycerol. Biophys J (1998) 74:1101–9. doi: 10.1016/S0006-3495(98)77828-2
18. King RJ, Carmichael MC, Horowitz PM. Reassembly of Lipid-Protein Complexes of Pulmonary Surfactant. Proposed Mechanism of Interaction. J Biol Chem (1983) 258:10672–80. doi: 10.1016/S0021-9258(17)44509-1
19. King RJ, Phillips MC, Horowitz PM, Dang SC. Interaction Between the 35 kDa Apolipoprotein of Pulmonary Surfactant and Saturated Phosphatidylcholines. Effects of temperature. Biochim Biophys Acta (1986) 879:1–13. doi: 10.1016/0005-2760(86)90259-6
20. Kuroki Y, Akino T. Pulmonary Surfactant Protein A (SP-A) Specifically Binds Dipalmitoylphosphatidylcholine. J Biol Chem (1991) 266:3068–73. doi: 10.1016/S0021-9258(18)49955-3
21. McCormack FX, Stewart J, Voelker DR, Damodarasamy M. Alanine Mutagenesis of Surfactant Protein A Reveals That Lipid Binding and pH-Dependent Liposome Aggregation Are Mediated by the Carbohydrate Recognition Domain. Biochemistry (1997) 36:13963–71. doi: 10.1021/bi970745q
22. Lopez-Rodriguez E, Pascual A, Arroyo R, Floros J, Perez-Gil J. Human Pulmonary Surfactant Protein SP-A1 Provides Maximal Efficiency of Lung Interfacial Films. Biophys J (2016) 111:524–36. doi: 10.1016/j.bpj.2016.06.025
23. Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in Biochemical Properties and in Biological Function Between Human SP-A1 and SP-A2 Variants, and the Impact of Ozone-Induced Oxidation. Biochemistry (2004) 43:4227–39. doi: 10.1021/bi036023i
24. Bruns G, Stroh H, Veldman GM, Latt SA, Floros J. The 35 Kd Pulmonary Surfactant-Associated Protein Is Encoded on Chromosome 10. Hum Genet (1987) 76:58–62. doi: 10.1007/BF00283051
25. Guo N, Mogues T, Weremowicz S, Morton CC, Sastry KN. The Human Ortholog of Rhesus Mannose-Binding Protein-A Gene Is an Expressed Pseudogene That Localizes to Chromosome 10. Mamm Genome (1998) 9:246–9. doi: 10.1007/s003359900735
26. Korfhagen TR, Glasser SW, Bruno MD, McMahan MJ, Whitsett JA. A Portion of the Human Surfactant Protein A (SP-A) Gene Locus Consists of a Pseudogene. Am J Respir Cell Mol Biol (1991) 4:463–9. doi: 10.1165/ajrcmb/4.5.463
27. Floros J, Wang G, Lin Z. Genetic Diversity ofa Molecule With Innate Host Defense and Surfactant-Related Functions; Characteristics, Primary Function, and Significance. Curr Pharmacogenomics (2005) 3:87–95. doi: 10.2174/1570160054022935
28. Karinch AM, Floros J. Translation In Vivo of 5’ Untranslated-Region Splice Variants of Human Surfactant Protein-a. Biochem J (1995) 307(Pt 2):327–30. doi: 10.1042/bj3070327
29. Karinch AM, Floros J. 5’ Splicing and Allelic Variants of the Human Pulmonary Surfactant Protein A Genes. Am J Respir Cell Mol Biol (1995) 12:77–88. doi: 10.1165/ajrcmb.12.1.7811473
30. Tsotakos N, Silveyra P, Lin Z, Thomas N, Vaid M, Floros J. Regulation of Translation by Upstream Translation Initiation Codons of Surfactant Protein A1 Splice Variants. Am J Physiol Lung Cell Mol Physiol (2015) 308:L58–75. doi: 10.1152/ajplung.00058.2014
31. Wang G, Guo X, Floros J. Differences in the Translation Efficiency and mRNA Stability Mediated by 5’-UTR Splice Variants of Human SP-A1 and SP-A2 Genes. Am J Physiol Lung Cell Mol Physiol (2005) 289:L497–508. doi: 10.1152/ajplung.00100.2005
32. Wang G, Guo X, Silveyra P, Kimball SR, Floros J. Cap-Independent Translation of Human SP-A 5’-UTR Variants: A Double-Loop Structure and Cis-Element Contribution. Am J Physiol Lung Cell Mol Physiol (2009) 296:L635–47. doi: 10.1152/ajplung.90508.2008
33. Wang G, Guo X, Floros J. Human SP-A 3’-UTR Variants Mediate Differential Gene Expression in Basal Levels and in Response to Dexamethasone. Am J Physiol Lung Cell Mol Physiol (2003) 284:L738–48. doi: 10.1152/ajplung.00375.2002
34. Hoover RR, Floros J. SP-A 3’-UTR Is Involved in the Glucocorticoid Inhibition of Human SP-A Gene Expression. Am J Physiol (1999) 276:L917–24. doi: 10.1152/ajplung.1999.276.6.L917
35. Floros J, Karinch AM, Human SP-A. Then and Now. Am J Physiol (1995) 268:L162–5. doi: 10.1152/ajplung.1995.268.2.L162
36. Floros J, DiAngelo S, Koptides M, Karinch AM, Rogan PK, Nielsen H, et al. Human SP-A Locus: Allele Frequencies and Linkage Disequilibrium Between the Two Surfactant Protein A Genes. Am J Respir Cell Mol Biol (1996) 15:489–98. doi: 10.1165/ajrcmb.15.4.8879183
37. Rishi A, Hatzis D, McAlmon K, Floros J. An Allelic Variant of the 6A Gene for Human Surfactant Protein a. Am J Physiol (1992) 262:L566–73. doi: 10.1152/ajplung.1992.262.5.L566
38. Voss T, Melchers K, Scheirle G, Schafer KP. Structural Comparison of Recombinant Pulmonary Surfactant Protein SP-A Derived From Two Human Coding Sequences: Implications for the Chain Composition of Natural Human SP-A. Am J Respir Cell Mol Biol (1991) 4:88–94. doi: 10.1165/ajrcmb/4.1.88
39. Phelps DS, Floros J. Localization of Surfactant Protein Synthesis in Human Lung by In Situ Hybridization. Am Rev Respir Dis (1988) 137:939–42. doi: 10.1164/ajrccm/137.4.939
40. Ochs M, Johnen G, Müller KM, Wahlers T, Hawgood S, Richter J, et al. Intracellular and Intraalveolar Localization of Surfactant Protein A (SP-A) in the Parenchymal Region of the Human Lung. Am J Respir Cell Mol Biol (2002) 26:91–8. doi: 10.1165/ajrcmb.26.1.4570
41. Chung J, Yu SH, Whitsett JA, Harding PG, Possmayer F. Effect of Surfactant-Associated Protein-A (SP-A) on the Activity of Lipid Extract Surfactant. Biochim Biophys Acta (1989) 1002:348–58. doi: 10.1016/0005-2760(89)90349-4
42. Li L, Zheng Q, Zhang Y, Li P, Fu Y, Hou J, et al. Antiviral Activity of Recombinant Porcine Surfactant Protein A Against Porcine Reproductive and Respiratory Syndrome Virus In Vitro. Arch Virol (2016) 161:1883–90. doi: 10.1007/s00705-016-2838-3
43. McCormack FX, Kuroki Y, Stewart JJ, Mason RJ, Voelker DR. Surfactant Protein A Amino Acids Glu195 and Arg197 Are Essential for Receptor Binding, Phospholipid Aggregation, Regulation of Secretion, and the Facilitated Uptake of Phospholipid by Type II Cells. J Biol Chem (1994) 269:29801–7. doi: 10.1016/S0021-9258(18)43952-X
44. Palaniyar N, Ridsdale RA, Hearn SA, Heng YM, Ottensmeyer FP, Possmayer F, et al. Filaments of Surfactant Protein A Specifically Interact With Corrugated Surfaces of Phospholipid Membranes. Am J Physiol (1999) 276:L631–41. doi: 10.1152/ajplung.1999.276.4.L631
45. Sun B, Curstedt T, Lindgren G, Franzén B, Alaiya AA, Calkovská A, et al. Biophysical and Physiological Properties of a Modified Porcine Surfactant Enriched With Surfactant Protein a. Eur Respir J (1997) 10:1967–74. doi: 10.1183/09031936.97.10091967
46. Terrasa AM, Guajardo MH, de Armas Sanabria E, Catalá A. Pulmonary Surfactant Protein A Inhibits the Lipid Peroxidation Stimulated by Linoleic Acid Hydroperoxide of Rat Lung Mitochondria and Microsomes. Biochim Biophys Acta (2005) 1735:101–10. doi: 10.1016/j.bbalip.2005.05.007
47. Worthman LA, Nag K, Rich N, Ruano ML, Casals C, Pérez-Gil J, et al. Pulmonary Surfactant Protein A Interacts With Gel-Like Regions in Monolayers of Pulmonary Surfactant Lipid Extract. Biophys J (2000) 79:2657–66. doi: 10.1016/S0006-3495(00)76504-0
48. Yamada T, Ikegami M, Tabor BL, Jobe AH. Effects of Surfactant Protein-A on Surfactant Function in Preterm Ventilated Rabbits. Am Rev Respir Dis (1990) 142:754–7. doi: 10.1164/ajrccm/142.4.754
49. Floros J, Wang G, Mikerov AN. Genetic Complexity of the Human Innate Host Defense Molecules, Surfactant Protein A1 (SP-A1) and SP-A2–impact on Function. Crit Rev eukaryotic Gene Expression (2009) 19:125–37. doi: 10.1615/CritRevEukarGeneExpr.v19.i2.30
50. Floros J. Human Surfactant Protein A (SP-A) Variants: Why So Many, Why Such a Complexity? Swiss Med Weekly (2001) 131:87–90.
51. Floros J, Wang G. A Point of View: Quantitative and Qualitative Imbalance in Disease Pathogenesis; Pulmonary Surfactant Protein A Genetic Variants as a Model. Comp Biochem Physiol Mol Integr Physiol (2001) 129:295–303. doi: 10.1016/S1095-6433(01)00325-7
52. Floros J, Phelps DS. Pulmonary Surfactant Protein A; Structure, Expression, and its Role in Innate Host Defense. In: Nakos G, L M, editors. Update of intensive care medicine. Ioannina, Greece (2002). p. 87–102. University of Ioannina.
53. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, et al. Novel, Non-Radioactive, Simple and Multiplex PCR-cRFLP Methods for Genotyping Human SP-A and SP-D Marker Alleles. Dis Markers (1999) 15:269–81. doi: 10.1155/1999/961430
54. Garcia-Verdugo I, Wang G, Floros J, Casals C. Structural Analysis and Lipid-Binding Properties of Recombinant Human Surfactant Protein a Derived From One or Both Genes. Biochemistry (2002) 41:14041–53. doi: 10.1021/bi026540l
55. Wang G, Myers C, Mikerov A, Floros J. Effect of Cysteine 85 on Biochemical Properties and Biological Function of Human Surfactant Protein A Variants. Biochemistry (2007) 46:8425–35. doi: 10.1021/bi7004569
56. Wang G, Guo X, Diangelo S, Thomas NJ, Floros J. Humanized SFTPA1 and SFTPA2 Transgenic Mice Reveal Functional Divergence of SP-A1 and SP-A2: Formation of Tubular Myelin In Vivo Requires Both Gene Products. J Biol Chem (2010) 285:11998–2010. doi: 10.1074/jbc.M109.046243
57. Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A Genetic Variants and Bleomycin-Induced Cytokine Production by THP-1 Cells: Effect of Ozone-Induced SP-A Oxidation. Am J Physiol Lung Cell Mol Physiol (2004) 286:L546–53. doi: 10.1152/ajplung.00267.2003
58. McCormack FX, Calvert HM, Watson PA, Smith DL, Mason RJ, Voelker DR. The Structure and Function of Surfactant Protein A Hydroxyproline- and Carbohydrate-Deficient Mutant Proteins. J Biol Chem (1994) 269:5833–41. doi: 10.1016/S0021-9258(17)37537-3
59. Phelps DS, Floros J. Proline Hydroxylation Alters the Electrophoretic Mobility of Pulmonary Surfactant-Associated Protein a. Electrophoresis (1988) 9:231–3. doi: 10.1002/elps.1150090508
60. Lamberg A, Helaakoski T, Myllyharju J, Peltonen S, Notbohm H, Pihlajaniemi T, et al. Characterization of Human Type III Collagen Expressed in a Baculovirus System. Production of a Protein with a Stable Triple Helix Requires Coexpression with the Two Types of Recombinant Prolyl 4-Hydroxylase Subunit. J Biol Chem (1996) 271:11988–95. doi: 10.1074/jbc.271.20.11988
61. Sanchez-Barbero F, Rivas G, Steinhilber W, Casals C. Structural and Functional Differences Among Human Surfactant Proteins SP-A1, SP-A2 and Co-Expressed SP-A1/SP-A2: Role of Supratrimeric Oligomerization. Biochem J (2007) 406:479–89. doi: 10.1042/BJ20070275
62. Chan VC, Ramshaw JA, Kirkpatrick A, Beck K, Brodsky B. Positional Preferences of Ionizable Residues in Gly-X-Y Triplets of the Collagen Triple-Helix. J Biol Chem (1997) 272:31441–6. doi: 10.1074/jbc.272.50.31441
63. Persikov AV, Ramshaw JA, Kirkpatrick A, Brodsky B. Peptide Investigations of Pairwise Interactions in the Collagen Triple-Helix. J Mol Biol (2002) 316:385–94. doi: 10.1006/jmbi.2001.5342
64. Selman M, Lin HM, Montano M, Jenkins AL, Estrada A, Lin Z, et al. And B Genetic Variants Predispose to Idiopathic Pulmonary Fibrosis. Hum Genet (2003) 113:542–50. doi: 10.1007/s00439-003-1015-4
65. Ikegami M, Elhalwagi BM, Palaniyar N, Dienger K, Korfhagen T, Whitsett JA, et al. The Collagen-Like Region of Surfactant Protein A (SP-A) Is Required for Correction of Surfactant Structural and Functional Defects in the SP-A Null Mouse. J Biol Chem (2001) 276:38542–8. doi: 10.1074/jbc.M102054200
66. Watson A, Kronqvist N, Spalluto CM, Griffiths M, Staples KJ, Wilkinson T, et al. Novel Expression of a Functional Trimeric Fragment of Human SP-A With Efficacy in Neutralisation of RSV. Immunobiology (2017) 222:111–8. doi: 10.1016/j.imbio.2016.10.015
67. deMello DE, Heyman S, Phelps DS, Floros J. Immunogold Localization of SP-A in Lungs of Infants Dying From Respiratory Distress Syndrome. Am J Pathol (1993) 142:1631–40.
68. Gómez-Gil L, Schürch D, Goormaghtigh E, Pérez-Gil J. Pulmonary Surfactant Protein SP-C Counteracts the Deleterious Effects of Cholesterol on the Activity of Surfactant Films Under Physiologically Relevant Compression-Expansion Dynamics. Biophys J (2009) 97:2736–45. doi: 10.1016/j.bpj.2009.08.045
69. Lukovic D, Cruz A, Gonzalez-Horta A, Almlen A, Curstedt T, Mingarro I, et al. Interfacial Behavior of Recombinant Forms of Human Pulmonary Surfactant Protein SP-C. Langmuir: ACS J surfaces colloids (2012) 28:7811–25. doi: 10.1021/la301134v
70. Paget TL, Parkinson-Lawrence EJ, Trim PJ, Autilio C, Panchal MH, Koster G, et al. Increased Alveolar Heparan Sulphate and Reduced Pulmonary Surfactant Amount and Function in the Mucopolysaccharidosis IIIA Mouse. Cells (2021) 10. doi: 10.3390/cells10040849
71. Serrano AG, Cruz A, Rodríguez-Capote K, Possmayer F, Pérez-Gil J. Intrinsic Structural and Functional Determinants Within the Amino Acid Sequence of Mature Pulmonary Surfactant Protein SP-B. Biochemistry (2005) 44:417–30. doi: 10.1021/bi048781u
72. Wang G, Taneva S, Keough KM, Floros J. Differential Effects of Human SP-A1 and SP-A2 Variants on Phospholipid Monolayers Containing Surfactant Protein B. Biochim Biophys Acta (2007) 1768:2060–9. doi: 10.1016/j.bbamem.2007.06.025
73. Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, et al. Characterization of a Human Surfactant Protein A1 (SP-A1) Gene-Specific Antibody; SP-A1 Content Variation Among Individuals of Varying Age and Pulmonary Health. Am J Physiol Lung Cell Mol Physiol (2007) 292:L1052–63. doi: 10.1152/ajplung.00249.2006
74. Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, et al. By Binding SIRPalpha or Calreticulin/CD91, Lung Collectins Act as Dual Function Surveillance Molecules to Suppress or Enhance Inflammation. Cell (2003) 115:13–23. doi: 10.1016/S0092-8674(03)00758-X
75. Geertsma MF, Nibbering PH, Haagsman HP, Daha MR, van Furth R. Binding of Surfactant Protein A to C1q Receptors Mediates Phagocytosis of Staphylococcus Aureus by Monocytes. Am J Physiol (1994) 267:L578–84. doi: 10.1152/ajplung.1994.267.5.L578
76. Kabha K, Schmegner J, Keisari Y, Parolis H, Schlepper-Schaeffer J, Ofek I. SP-A Enhances Phagocytosis of Klebsiella by Interaction With Capsular Polysaccharides and Alveolar Macrophages. Am J Physiol (1997) 272:L344–52. doi: 10.1152/ajplung.1997.272.2.L344
77. Sever-Chroneos Z, Krupa A, Davis J, Hasan M, Yang CH, Szeliga J, et al. (SP-A)-Mediated Clearance of Staphylococcus Aureus Involves Binding of SP-A to the Staphylococcal Adhesin Eap and the Macrophage Receptors SP-A Receptor 210 and Scavenger Receptor Class a. J Biol Chem (2011) 286:4854–70. doi: 10.1074/jbc.M110.125567
78. Mikerov AN, Umstead TM, Huang W, Liu W, Phelps DS, Floros J. SP-A1 and SP-A2 Variants Differentially Enhance Association of Pseudomonas Aeruginosa With Rat Alveolar Macrophages. Am J Physiol Lung Cell Mol Physiol (2005) 288:L150–8. doi: 10.1152/ajplung.00135.2004
79. Mikerov AN, Wang G, Umstead TM, Zacharatos M, Thomas NJ, Phelps DS, et al. Surfactant Protein A2 (SP-A2) Variants Expressed in CHO Cells Stimulate Phagocytosis of Pseudomonas Aeruginosa More Than do SP-A1 Variants. Infection Immun (2007) 75:1403–12. doi: 10.1128/IAI.01341-06
80. Floros J, Steinbrink R, Jacobs K, Phelps D, Kriz R, Recny M, et al. Isolation and Characterization of cDNA Clones for the 35-kDa Pulmonary Surfactant-Associated Protein. J Biol Chem (1986) 261:9029–33. doi: 10.1016/S0021-9258(19)84483-6
81. Phelps DS, Floros J, Taeusch HW Jr. Post-Translational Modification of the Major Human Surfactant-Associated Proteins. Biochem J (1986) 237:373–7. doi: 10.1042/bj2370373
82. Haagsman HP, Diemel RV. Surfactant-Associated Proteins: Functions and Structural Variation. Comp Biochem Physiol Mol Integr Physiol (2001) 129:91–108. doi: 10.1016/S1095-6433(01)00308-7
83. Khubchandani KR, Goss KL, Engelhardt JF, Snyder JM. In Situ Hybridization of SP-A mRNA in Adult Human Conducting Airways. Pediatr Pathol Mol Med (2001) 20:349–66. doi: 10.1080/15513810109168620
84. Elhalwagi BM, Damodarasamy M, McCormack FX. Alternate Amino Terminal Processing of Surfactant Protein A Results in Cysteinyl Isoforms Required for Multimer Formation. Biochemistry (1997) 36:7018–25. doi: 10.1021/bi970100q
85. Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, et al. Impact of Ozone Exposure on the Phagocytic Activity of Human Surfactant Protein A (SP-A) and SP-A Variants. Am J Physiol Lung Cell Mol Physiol (2008) 294:L121–30. doi: 10.1152/ajplung.00288.2007
86. Oberley RE, Snyder JM. Recombinant Human SP-A1 and SP-A2 Proteins Have Different Carbohydrate-Binding Characteristics. Am J Physiol Lung Cell Mol Physiol (2003) 284:L871–81. doi: 10.1152/ajplung.00241.2002
87. Wang G, Phelps DS, Umstead TM, Floros J. Human SP-A Protein Variants Derived From One or Both Genes Stimulate TNF-Alpha Production in the THP-1 Cell Line. Am J Physiol Lung Cell Mol Physiol (2000) 278:L946–54. doi: 10.1152/ajplung.2000.278.5.L946
88. Wang G, Umstead TM, Phelps DS, Al-Mondhiry H, Floros J. The Effect of Ozone Exposure on the Ability of Human Surfactant Protein a Variants to Stimulate Cytokine Production. Environ Health Perspect (2002) 110:79–84. doi: 10.1289/ehp.0211079
89. Umstead TM, Phelps DS, Wang G, Floros J, Tarkington BK. In Vitro Exposure of Proteins to Ozone. Toxicol Mech Methods (2002) 12:1–16. doi: 10.1080/15376510209167932
90. Oosting RS, van Greevenbroek MM, Verhoef J, van Golde LM, Haagsman HP. Structural and Functional Changes of Surfactant Protein A Induced by Ozone. Am J Physiol (1991) 261:L77–83. doi: 10.1152/ajplung.1991.261.2.L77
91. Oosting RS, Van Iwaarden JF, Van Bree L, Verhoef J, Van Golde LM, Haagsman HP. Exposure of Surfactant Protein A to Ozone In Vitro and In Vivo Impairs Its Interactions With Alveolar Cells. Am J Physiol (1992) 262:L63–8. doi: 10.1152/ajplung.1992.262.1.L63
92. Mikerov AN, White M, Hartshorn K, Wang G, Floros J. Inhibition of Hemagglutination Activity of Influenza A Viruses by SP-A1 and SP-A2 Variants Expressed in CHO Cells. Med Microbiol Immunol (2008) 197:9–12. doi: 10.1007/s00430-007-0051-4
93. Arnon TI, Achdout H, Lieberman N, Gazit R, Gonen-Gross T, Katz G, et al. The Mechanisms Controlling the Recognition of Tumor- and Virus-Infected Cells by Nkp46. Blood (2004) 103:664–72. doi: 10.1182/blood-2003-05-1716
94. Lee EU, Roth J, Paulson JC. Alteration of Terminal Glycosylation Sequences on N-Linked Oligosaccharides of Chinese Hamster Ovary Cells by Expression of Beta-Galactoside Alpha 2,6-Sialyltransferase. J Biol Chem (1989) 264:13848–55. doi: 10.1016/S0021-9258(18)80078-3
95. Herrera-Ramos E, López-Rodríguez M, Ruíz-Hernández JJ, Horcajada JP, Borderías L, Lerma E, et al. Surfactant Protein A Genetic Variants Associate With Severe Respiratory Insufficiency in Pandemic Influenza A Virus Infection. Crit Care (London England) (2014) 18:R127. doi: 10.1186/cc13934
96. Pederson WP, Cyphert-Daly JM, Tighe RM, Que LG, Ledford JG. Genetic Variation in Surfactant Protein-A2 Alters Responses to Ozone. PloS One (2021) 16:e0247504. doi: 10.1371/journal.pone.0247504
97. Ali M, Umstead TM, Haque R, Mikerov AN, Freeman WM, Floros J, et al. Differences in the BAL Proteome After Klebsiella Pneumoniae Infection in Wild Type and SP-A-/- Mice. Proteome Sci (2010) 8:34. doi: 10.1186/1477-5956-8-34
98. Cogen AL, Moore TA. Beta2-Microglobulin-Dependent Bacterial Clearance and Survival During Murine Klebsiella Pneumoniae Bacteremia. Infection Immun (2009) 77:360–6. doi: 10.1128/IAI.00909-08
99. Markart P, Korfhagen TR, Weaver TE, Akinbi HT. Mouse Lysozyme M Is Important in Pulmonary Host Defense Against Klebsiella Pneumoniae Infection. Am J Respir Crit Care Med (2004) 169:454–8. doi: 10.1164/rccm.200305-669OC
100. Mikerov AN, Haque R, Gan X, Guo X, Phelps DS, Floros J. Ablation of SP-A has a Negative Impact on the Susceptibility of Mice to Klebsiella Pneumoniae Infection After Ozone Exposure: Sex Differences. Respir Res (2008) 9:77. doi: 10.1186/1465-9921-9-77
101. Haque R, Umstead TM, Ahn K, Phelps DS, Floros J. Effect of Low Doses of Lipopolysaccharide Prior to Ozone Exposure on Bronchoalveolar Lavage: Differences Between Wild Type and Surfactant Protein A-Deficient Mice. Pneumon (2009) 22:143–55.
102. Haque R, Umstead TM, Freeman WM, Floros J, Phelps DS. The Impact of Surfactant Protein-A on Ozone-Induced Changes in the Mouse Bronchoalveolar Lavage Proteome. Proteome Sci (2009) 7:12. doi: 10.1186/1477-5956-7-12
103. Xu HN, Lin Z, Gandhi CK, Amatya S, Wang Y, Lin LZ, et al. Sex and SP-A2 Dependent NAD(H) Redox Alterations in Mouse Alveolar Macrophages in Response to Ozone Exposure: Potential Implications for COVID-19. Antioxidants (accepted; press) (2020). doi: 10.3390/antiox9100915
104. Phelps DS, Umstead TM, Floros J. Sex Differences in the Response of the Alveolar Macrophage Proteome to Treatment With Exogenous Surfactant Protein-a. Proteome Sci (2012) 10:44. doi: 10.1186/1477-5956-10-44
105. Phelps DS, Umstead TM, Quintero OA, Yengo CM, Floros J. In Vivo Rescue of Alveolar Macrophages From SP-A Knockout Mice With Exogenous SP-A Nearly Restores a Wild Type Intracellular Proteome; Actin Involvement. Proteome Sci (2011) 9:67. doi: 10.1186/1477-5956-9-67
106. Abbondanza C, Rossi V, Roscigno A, Gallo L, Belsito A, Piluso G, et al. Interaction of Vault Particles With Estrogen Receptor in the MCF-7 Breast Cancer Cell. J Cell Biol (1998) 141:1301–10. doi: 10.1083/jcb.141.6.1301
107. El Marzouk S, Schultz-Norton JR, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Rho GDP Dissociation Inhibitor Alpha Interacts With Estrogen Receptor Alpha and Influences Estrogen Responsiveness. J Mol Endocrinol (2007) 39:249–59. doi: 10.1677/JME-07-0055
108. Klimczak M, Biecek P, Zylicz A, Zylicz M. Heat Shock Proteins Create a Signature to Predict the Clinical Outcome in Breast Cancer. Sci Rep (2019) 9:7507. doi: 10.1038/s41598-019-43556-1
109. Durrani F, Phelps DS, Weisz J, Silveyra P, Hu S, Mikerov AN, et al. Gonadal Hormones and Oxidative Stress Interaction Differentially Affects Survival of Male and Female Mice After Lung Klebsiella Pneumoniae Infection. Exp Lung Res (2012) 38:165–72. doi: 10.3109/01902148.2011.654045
110. Phelps DS, Umstead TM, Quintero OA, Floros J, Surfactant protein A. (SP-A)1 and SP-A2 Differentially Affect F-Actin Levels in the Alveolar Macrophage. AJBSR (2021). doi: 10.34297/AJBSR.2021.12.001740
111. Wang G, Umstead TM, Hu S, Mikerov AN, Phelps DS, Floros J. Differential Effects of Human SP-A1 and SP-A2 on the BAL Proteome and Signaling Pathways in Response to Klebsiella Pneumoniae and Ozone Exposure. Front Immunol (2019) 10:561. doi: 10.3389/fimmu.2019.00561
112. Phelps DS, Umstead TM, Silveyra P, Hu S, Wang G, Floros J. Differences in the Alveolar Macrophage Proteome in Transgenic Mice Expressing Human SP-A1 and SP-A2. J Proteomics Genomics Res (2013) 1:2–26. doi: 10.14302/issn.2326-0793.jpgr-12-207
113. Nalian A, Umstead TM, Yang CH, Silveyra P, Thomas NJ, Floros J, et al. Structural and Functional Determinants of Rodent and Human Surfactant Protein A: A Synthesis of Binding and Computational Data. Front Immunol (2019) 10:2613. doi: 10.3389/fimmu.2019.02613
114. Phelps DS, Umstead TM, Floros J. Sex Differences in the Acute In Vivo Effects of Different Human SP-A Variants on the Mouse Alveolar Macrophage Proteome. J Proteomics (2014) 108:427–44. doi: 10.1016/j.jprot.2014.06.007
115. Tino MJ, Wright JR. Surfactant proteins A And D Specifically Stimulate Directed Actin-Based Responses in Alveolar Macrophages. Am J Physiol (1999) 276:L164–74. doi: 10.1152/ajplung.1999.276.1.L164
116. Tsotakos N, Phelps DS, Yengo CM, Chinchilli VM, Floros J. Single-Cell Analysis Reveals Differential Regulation of the Alveolar Macrophage Actin Cytoskeleton by Surfactant Proteins A1 and A2: Implications of Sex and Aging. Biol sex Dif (2016) 7:18. doi: 10.1186/s13293-016-0071-0
117. Thorenoor N, Kawasawa YI, Gandhi CK, Zhang X, Floros J. Differential Impact of Co-Expressed SP-A1/SP-A2 Protein on AM Mirnome; Sex Differences. Front Immunol (2019) 10:1960. doi: 10.3389/fimmu.2019.01960
118. Thorenoor N, Umstead TM, Zhang X, Phelps DS, Floros J. Survival of Surfactant Protein-A1 and SP-A2 Transgenic Mice After Klebsiella Pneumoniae Infection, Exhibits Sex-, Gene-, and Variant Specific Differences; Treatment With Surfactant Protein Improves Survival. Front Immunol (2018) 9:2404. doi: 10.3389/fimmu.2018.02404
119. Liu Y, Tejpal N, You J, Li XC, Ghobrial RM, Kloc M. ROCK Inhibition Impedes Macrophage Polarity and Functions. Cell Immunol (2016) 300:54–62. doi: 10.1016/j.cellimm.2015.12.005
120. Lafuse WP, Rajaram MVS, Wu Q, Moliva JI, Torrelles JB, Turner J, et al. Identification of an Increased Alveolar Macrophage Subpopulation in Old Mice That Displays Unique Inflammatory Characteristics and Is Permissive to Mycobacterium Tuberculosis Infection. J Immunol (2019) 203:2252–64. doi: 10.4049/jimmunol.1900495
121. Svedberg FR, Brown SL, Krauss MZ, Campbell L, Sharpe C, Clausen M, et al. The Lung Environment Controls Alveolar Macrophage Metabolism and Responsiveness in Type 2 Inflammation. Nat Immunol (2019) 20:571–80. doi: 10.1038/s41590-019-0352-y
122. Noutsios GT, Thorenoor N, Zhang X, Phelps DS, Umstead TM, Durrani F, et al. Major Effect of Oxidative Stress on the Male, But Not Female, SP-A1 Type II Cell Mirnome. Front Immunol (2019) 10:1514. doi: 10.3389/fimmu.2019.01514
123. Noutsios GT, Thorenoor N, Zhang X, Phelps DS, Umstead TM, Durrani F, et al. SP-A2 Contributes to miRNA-Mediated Sex Differences in Response to Oxidative Stress: Pro-Inflammatory, Anti-Apoptotic, and Anti-Oxidant Pathways Are Involved. Biol sex Dif (2017) 8:37. doi: 10.1186/s13293-017-0158-2
124. Taneja V. Sex Hormones Determine Immune Response. Front Immunol (2018) 9:1931. doi: 10.3389/fimmu.2018.01931
125. Thorenoor N, Phelps DS, Floros J. Differential Sex-Dependent Regulation of the Alveolar Macrophage Mirnome of SP-A2 and Co-Ex (SP-A1/SP-A2) and Sex Differences Attenuation After 18 H of Ozone Exposure. Antioxidants (Basel Switzerland) (2020) 9. doi: 10.3390/antiox9121190
126. Thorenoor N, Kawasawa YI, Gandhi CK, Floros J. Sex-Specific Regulation of Gene Expression Networks by Surfactant Protein A (SP-A) Variants in Alveolar Macrophages in Response to Klebsiella Pneumoniae. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.01290
127. Schubert W. Exploring Molecular Networks Directly in the Cell. Cytometry Part A: J Int Soc Analytical Cytol (2006) 69:109–12. doi: 10.1002/cyto.a.20234
128. Schubert W. A Three-Symbol Code for Organized Proteomes Based on Cyclical Imaging of Protein Locations. Cytometry Part A: J Int Soc Analytical Cytol (2007) 71:352–60. doi: 10.1002/cyto.a.20281
129. Schubert W. Systematic, Spatial Imaging of Large Multimolecular Assemblies and the Emerging Principles of Supramolecular Order in Biological Systems. J Mol Recognition: JMR (2014) 27:3–18. doi: 10.1002/jmr.2326
130. Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Böckelmann R, Malykh Y, et al. Analyzing Proteome Topology and Function by Automated Multidimensional Fluorescence Microscopy. Nat Biotechnol (2006) 24:1270–8. doi: 10.1038/nbt1250
131. Schubert W, Gieseler A, Krusche A, Serocka P, Hillert R. Next-Generation Biomarkers Based on 100-Parameter Functional Super-Resolution Microscopy TIS. New Biotechnol (2012) 29:599–610. doi: 10.1016/j.nbt.2011.12.004
132. Phelps DS, Chinchilli VM, Weisz J, Yang L, Shearer D, Zhang X, et al. Differences in the Alveolar Macrophage Toponome in Humanized SP-A1 and SP-A2 Transgenic Mice. JCI Insight (2020) 5. doi: 10.1172/jci.insight.141410
133. Bhattacharya S, Mathew G, Ruban E, Epstein DB, Krusche A, Hillert R, et al. Toponome Imaging System: In Situ Protein Network Mapping in Normal and Cancerous Colon From the Same Patient Reveals More Than Five-Thousand Cancer Specific Protein Clusters and Their Subcellular Annotation by Using a Three Symbol Code. J Proteome Res (2010) 9:6112–25. doi: 10.1021/pr100157p
134. Bode M, Irmler M, Friedenberger M, May C, Jung K, Stephan C, et al. Interlocking transcriptomics, Proteomics and Toponomics Technologies for Brain Tissue Analysis in Murine Hippocampus. Proteomics (2008) 8:1170–8. doi: 10.1002/pmic.200700742
135. Phelps DS, Chinchilli VM, Weisz J, Shearer D, Zhang X, Floros J. Using Toponomics to Characterize Phenotypic Diversity in Alveolar Macrophages From Male Mice Treated With Exogenous SP-A1. Biomarker Res (2020) 8:5. doi: 10.1186/s40364-019-0181-z
136. Thorenoor N, Zhang X, Umstead TM, Scott Halstead E, Phelps DS, Floros J. Differential Effects of Innate Immune Variants of Surfactant Protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in Airway Function After Klebsiella Pneumoniae Infection and Sex Differences. Respir Res (2018) 19:23. doi: 10.1186/s12931-018-0723-1
137. Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, et al. Surfactant Protein A is Defective in Abrogating Inflammation in Asthma. Am J Physiol Lung Cell Mol Physiol (2011) 301:L598–606. doi: 10.1152/ajplung.00381.2010
138. Kremlev SG, Umstead TM, Phelps DS. Surfactant Protein A Regulates Cytokine Production in the Monocytic Cell Line THP-1. Am J Physiol (1997) 272:L996–1004. doi: 10.1152/ajplung.1997.272.5.L996
139. Thorenoor N, Phelps DS, Kala P, Ravi R, Floros Phelps A, Umstead TM, et al. Impact of Surfactant Protein-A Variants on Survival in Aged Mice in Response to Klebsiella pneumoniae Infection and Ozone: Serendipity in Action. Microorganisms (2020) 8. doi: 10.3390/microorganisms8091276
140. Abbasi A, Phelps DS, Ravi R, Floros J. Can Prophylactic High Flow of Humidified and Warmed Filtered Air Improve Survival From Bacterial Pneumonia and SARS-CoV-2 in Elderly Individuals? The Role of Surfactant Protein a. Antioxidants (Basel Switzerland) (2021) 10. doi: 10.3390/antiox10050640
141. D’Ovidio F, Floros J, Aramini B, Lederer D, DiAngelo SL, Arcasoy S, et al. Donor Surfactant Protein A2 Polymorphism and Lung Transplant Survival. Eur Respir J (2019). doi: 10.1183/13993003.00618-2019
142. Aramini B, Geraghty P, Lederer DJ, Costa J, DiAngelo SL, Floros J, et al. And D Polymorphisms and Methylprednisolone Pharmacogenetics in Donor Lungs. J thoracic Cardiovasc Surg (2019) 157:2109–17. doi: 10.1016/j.jtcvs.2018.12.098
143. D’Ovidio F, Kaneda H, Chaparro C, Mura M, Lederer D, Di Angelo S, et al. Pilot Study Exploring Lung Allograft Surfactant Protein A (SP-A) Expression in Association With Lung Transplant Outcome. Am J Transplantation: Off J Am Soc Transplant Am Soc Transplant Surgeons (2013) 13:2722–9. doi: 10.1111/ajt.12407
144. Floros J, Fan R, Surfactant protein A. And B Genetic Variants and Respiratory Distress Syndrome: Allele Interactions. Biol Neonate (2001) 80(Suppl 1):22–5. doi: 10.1159/000047173
145. Floros J, Kala P. Surfactant Proteins: Molecular Genetics of Neonatal Pulmonary Diseases. Annu Rev Physiol (1998) 60:365–84. doi: 10.1146/annurev.physiol.60.1.365
146. Silveyra P, Floros J. Genetic Variant Associations of Human SP-A and SP-D With Acute and Chronic Lung Injury. Front Biosci: J Virtual Library (2012) 17:407–29. doi: 10.2741/3935
147. Floros J, Thomas NJ. Genetic Variations of Surfactant Proteins and Lung Injury. In: Nakos G, Papathanasiou A, editors. In Surfactant in Pathogenesis and Treatment of Lung Disease. Kerala, India: Research Signpost (2009). p. 25–48.
148. Lin Z, Thorenoor N, Wu R, DiAngelo SL, Ye M, Thomas NJ, et al. Genetic Association of Pulmonary Surfactant Protein Genes, SFTPA1, SFTPA2and SFTPD With Cystic Fibrosis. Front Immunol (2018) 9. doi: 10.3389/fimmu.2018.02256
149. D’Ovidio F, Floros J, Aramini B, Lederer D, DiAngelo SL, Arcasoy S, et al. Donor Surfactant Protein A2 Polymorphism and Lung Transplant Survival. Eur Respir J (2020) 55. doi: 10.1183/13993003.00618-2019
150. Gandhi CK, Chen C, Wu R, Yang L, Thorenoor N, Thomas NJ, et al. Association of SNP-SNP Interactions of Surfactant Protein Genes With Pediatric Acute Respiratory Failure. J Clin Med (2020) 9. doi: 10.3390/jcm9041183
151. Gandhi CK, Chen C, Amatya S, Yang L, Fu C, Zhou S, et al. SNP and Haplotype Interaction Models Reveal Association of Surfactant Protein Gene Polymorphisms With Hypersensitivity Pneumonitis of Mexican Population. Front Med (2020) 7:588404. doi: 10.3389/fmed.2020.588404
152. Nathan N, Giraud V, Picard C, Nunes H, Dastot-Le Moal F, Copin B, et al. Germline SFTPA1 Mutation in Familial Idiopathic Interstitial Pneumonia and Lung Cancer. Hum Mol Genet (2016) 25:1457–67. doi: 10.1183/13993003.congress-2016.OA481
153. Tsitoura MI, Stavrou EF, Maraziotis IA, Sarafidis K, Athanassiadou A, Dimitriou G, et al. And B Gene Polymorphisms and Risk of Respiratory Distress Syndrome in Late-Preterm Neonates. PloS One (2016) 11:e0166516. doi: 10.1371/journal.pone.0166516
154. Floros J, Phelps DS. Is the Role of Lung Innate Immune Molecules, SP-A1 and SP-A2, and of the Alveolar Macrophage Being Overlooked in COVID-19 Diverse Outcomes? PNEUMON (2020) 33:1–5.
155. Tekos F, Skaperda Z, Goutzourelas N, Phelps DS, Floros J, Kouretas D. The Importance of Redox Status in the Frame of Lifestyle Approaches and the Genetics of the Lung Innate Immune Molecules, SP-A1 and SP-A2, on Differential Outcomes of COVID-19 Infection. Antioxidants (Basel Switzerland) (2020) 9. doi: 10.3390/antiox9090784
156. Mikerov AN, Hu S, Durrani F, Gan X, Wang G, Umstead TM, et al. Impact of Sex and Ozone Exposure on the Course of Pneumonia in Wild(-/-) Mice. Microbial Pathogenesis (2012) 52:239–49. doi: 10.1016/j.micpath.2012.01.005
157. LeVine AM, Gwozdz J, Stark J, Bruno M, Whitsett J, Korfhagen T. Surfactant Protein-A Enhances Respiratory Syncytial Virus Clearance In Vivo. J Clin Invest (1999) 103:1015–21. doi: 10.1172/JCI5849
158. LeVine AM, Hartshorn K, Elliott J, Whitsett J, Korfhagen T. Absence of SP-A Modulates Innate and Adaptive Defense Responses to Pulmonary Influenza Infection. Am J Physiol Lung Cell Mol Physiol (2002) 282:L563–72. doi: 10.1152/ajplung.00280.2001
159. Harrod KS, Trapnell BC, Otake K, Korfhagen TR, Whitsett JA. SP-A Enhances Viral Clearance and Inhibits Inflammation After Pulmonary Adenoviral Infection. Am J Physiol (1999) 277:L580–8. doi: 10.1152/ajplung.1999.277.3.L580
160. El Saleeby CM, Li R, Somes GW, Dahmer MK, Quasney MW, DeVincenzo JP. Surfactant Protein A2 Polymorphisms and Disease Severity in a Respiratory Syncytial Virus-Infected Population. J Pediatr (2010) 156:409–14. doi: 10.1016/j.jpeds.2009.09.043
161. Lofgren J, Ramet M, Renko M, Marttila R, Hallman M. Association Between Surfactant Protein A Gene Locus and Severe Respiratory Syncytial Virus Infection in Infants. J Infect Dis (2002) 185:283–9. doi: 10.1086/338473
162. Silveyra P, Floros J. Genetic Complexity of the Human Surfactant-Associated Proteins SP-A1 and SP-A2. Gene (2013) 531:126–32. doi: 10.1016/j.gene.2012.09.111
163. Floros J, Thomas NJ. Surfactant Protein Genetics in Community-Acquired Pneumonia: Balancing the Host Inflammatory State. Crit Care (London England) (2011) 15:156. doi: 10.1186/cc10115
164. Floros J. Why Two Surfactant Protein A Genes in Human? Are These Functionally and Structurally Equivalent? Appl Cardiopulmonary Pathophysiol (2004) 13:32–4.
165. Floros J, Phelps D. Human Surfactant Proteins (SP-) A1 and SP-A2: To Include or Not in Surfactant Replacement Therapy, and If Yes, Both or Which One? J Neonatal Biol (2014) 3. doi: 10.4172/2167-0897.1000136
166. Avery ME, Mead J. Surface Properties in Relation to Atelectasis and Hyaline Membrane Disease. Am J Dis Child (1959) 97:517–23. doi: 10.1001/archpedi.1959.02070010519001
167. deMello DE, Phelps DS, Patel G, Floros J, Lagunoff D. Expression of the 35kda and Low Molecular Weight Surfactant-Associated Proteins in the Lungs of Infants Dying With Respiratory Distress Syndrome. Am J Pathol (1989) 134:1285–93.
168. Stevens PA, Schadow B, Bartholain S, Segerer H, Obladen M. Surfactant Protein A in the Course of Respiratory Distress Syndrome. Eur J Pediatr (1992) 151:596–600. doi: 10.1007/BF01957730
169. Noutsios GT, Floros J. Highlights of Early Pulmonary Surfactant: Research From Bench to Clinic. Pneumon (2013) 26(4):350–4.
Keywords: alveolar macrophage, SFTPA1, SFTPA2, SP-A1, SP-A2
Citation: Floros J, Thorenoor N, Tsotakos N and Phelps DS (2021) Human Surfactant Protein SP-A1 and SP-A2 Variants Differentially Affect the Alveolar Microenvironment, Surfactant Structure, Regulation and Function of the Alveolar Macrophage, and Animal and Human Survival Under Various Conditions. Front. Immunol. 12:681639. doi: 10.3389/fimmu.2021.681639
Received: 16 March 2021; Accepted: 02 July 2021;
Published: 17 August 2021.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Taruna Madan, National Institute for Research in Reproductive Health (ICMR), IndiaAnthony George Tsolaki, Brunel University London, United Kingdom
Raymond B Birge, Rutgers, The State University of New Jersey, United States
Copyright © 2021 Floros, Thorenoor, Tsotakos and Phelps. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Floros, jfloros@psu.edu
 Joanna Floros
Joanna Floros Nithyananda Thorenoor
Nithyananda Thorenoor Nikolaos Tsotakos
Nikolaos Tsotakos David S. Phelps1
David S. Phelps1
