Abstract
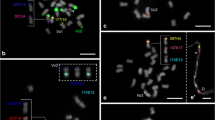
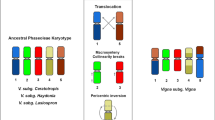
Comparative cytogenetic mapping is a powerful approach to gain insights into genome organization of orphan crops, lacking a whole sequenced genome. To investigate the cytogenomic evolution of important Vigna and Phaseolus beans, we built a BAC-FISH (fluorescent in situ hybridization of bacterial artificial chromosome) map of Vigna aconitifolia (Vac, subgenus Ceratotropis), species with no sequenced genome, and compared with V. unguiculata (Vu, subgenus Vigna) and Phaseolus vulgaris (Pv) maps. Seventeen Pv BACs, eight Vu BACs, and 5S and 35S rDNA probes were hybridized in situ on the 11 Vac chromosome pairs. Five Vac chromosomes (Vac6, Vac7, Vac9, Vac10, and Vac11) showed conserved macrosynteny and collinearity between V. unguiculata and P. vulgaris. On the other hand, we observed collinearity breaks, identified by pericentric inversions involving Vac2 (Vu2), Vac4 (Vu4), and Vac3 (Pv3). We also detected macrosynteny breaks of translocation type involving chromosomes 1 and 8 of V. aconitifolia and P. vulgaris; 2 and 3 of V. aconitifolia and P. vulgaris; and 1 and 5 of V. aconitifolia and V. unguiculata. Considering our data and previous BAC-FISH studies, six chromosomes (1, 2, 3, 4, 5, and 8) are involved in major karyotype divergences between genera and five (1, 2, 3, 4, and 5) between Vigna subgenera, including mechanisms such as duplications, inversions, and translocations. Macrosynteny breaks between Vigna and Phaseolus suggest that the major chromosomal rearrangements have occurred within the Vigna clade. Our cytogenomic comparisons bring new light on the degree of shared macrosynteny and mechanisms of karyotype diversification during Vigna and Phaseolus evolution.




Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- Chr:
-
Chromosome
- DAPI:
-
4,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- LG:
-
Linkage group
- Mya:
-
Million years ago
- rDNA:
-
Ribosomal DNA
- Pl :
-
Phaseolus lunatus
- Pm :
-
Phaseolus microcarpus
- Pv :
-
Phaseolus vulgaris
- Vac :
-
Vigna aconitifolia
- Vm :
-
Vigna mungo
- Vum :
-
Vigna umbellata
- Vu :
-
Vigna unguiculata
References
Almeida C, Pedrosa-Harand A (2011) Contrasting rDNA evolution in lima bean (Phaseolus lunatus L.) and common bean (P. vulgaris L. Fabaceae). Cytogenet Genome Res 132:212–217. https://doi.org/10.1159/000321677
Almeida C, Pedrosa-Harand A (2013) High macro-collinearity between lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.) as revealed by comparative cytogenetic mapping. Theor Appl Genet 126:1909–1916. https://doi.org/10.1007/s00122-013-2106-9
Bonifácio EM, Fonsêca A, Almeida C, Santos KGB, Pedrosa-Harand A (2012) Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124:1513–1520. https://doi.org/10.1007/s00122-012-1806-x
Brink M, Jansen PCM (2006) Vigna aconitifolia (Jacq.) Maréchal. In: Brink M, Belay G (eds) PROTA 1: cereals and pulses/Céréales et Légumes Secs. Wageningen, Netherlands, pp 1–5
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.): model food legumes. Plant Soil 252:55–128. https://doi.org/10.1023/A:1024146710611
Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
Chaitieng B, Kaga A, Tomooka N, Isemura T, Kuroda Y, Vaughan DA (2006) Development of a black gram [Vigna mungo (L.) Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Wild.) Ohwi and Ohashi] linkage map. Theor Appl Genet 113:1261–1269. https://doi.org/10.1007/s00122-006-0380-5
Chang Y, Liu H, Liu M, Liao X, Sahu SK, Fu Y, Song B, Cheng S, Kariba R, Muthemba S, Hendre PS, Mayes S, Ho WK, Yssel AEJ, Kendabie P, Wang S, Li L, Muchugi A, Jamnadass R, Lu H, Peng S, van Deynze A, Simons A, Yana-Shapiro H, van de Peer Y, Xu X, Yang H, Wang J, Liu X (2019) The draft genomes of five agriculturally important African orphan crops. GigaScience 8:1–16. https://doi.org/10.1093/gigascience/giy152
Delgado-Salinas A, Thulin M, Pasquet R, Weeden LM (2011) Vigna (Leguminosae) sensu lato: the names and identities of the American segregate genera. Am J Bot 98:1694–1715. https://doi.org/10.3732/ajb.1100069
Fonsêca AFA, Pedrosa-Harand A (2013) Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 56:335–343. https://doi.org/10.1139/gen-2013-0025
Fonsêca AFA, Ferreira J, Santos TRB et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Fonsêca AFA, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma. 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Forni-Martins ER (1986) New chromosome number in the genus Vigna Savi (Leguminosae-Papilionoideae). Bull Jardin Bot Nat Belgique 56:129–133
Freyre R, Skroch PW, Geffroy V, Adam-Blondon AF, Shirmohamadali A, Johnson WC, Llaca V, Nodari RO, Pereira PA, Tsai SM, Tohme J, Dron M, Nienhuis J, Vallejos CE, Gepts P (1998) Towards an integrated linkage map of common bean 4: development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97:847–856. https://doi.org/10.1007/s001220050964
Gepts P, Aragão FJL, Barros E et al (2008) Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Moore PH, Ming R (eds) Genomics of tropical crop plants. Springer, New York, pp 113–143
Gupta SK, Souframanien J, Gopalakrishna T (2008) Construction of a genetic linkage map of black gram, Vigna mungo (L.) Hepper, based on molecular markers and comparative studies. Genome 51:628–637. https://doi.org/10.1139/G08-050
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373. https://doi.org/10.1016/0168-9525(92)90287-E
Hougaard BK, Madsen LH, Sandal N, de Carvalho Moretzsohn M, Fredslund J, Schauser L, Nielsen AM, Rohde T, Sato S, Tabata S, Bertioli DJ, Stougaard J (2008) Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics 179:2299–2312. https://doi.org/10.1534/genetics.108.090084
Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA (2010) The genetics of domestication of rice bean, Vigna umbellata. Ann Bot 106:927–944. https://doi.org/10.1093/aob/mcq188
Iwata A, Greenland CM, Jackson AS (2013) Cytogenetics of legumes in the Phaseoloid clade. Plant Genome 6:1–8. https://doi.org/10.3835/plantgenome2013.03.0004
Iwata-Otsubo A, Jer-Young L, Gill N, Jackson SA (2016) Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosom Res 24:197–216. https://doi.org/10.1007/s10577-015-9515-3
Javadi F, Tun YT, Kawase M, Guan K, Yamaguchi H (2011) Molecular phylogeny of the subgenus Ceratotropis (genus Vigna, Leguminosae) reveals three eco-geographical groups and Late Pliocene-Pleistocene diversification: evidence from four plastid DNA region sequences. Ann Bot 8:367–380. https://doi.org/10.1093/aob/mcr141
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. PNAS 105:5833–5838. https://doi.org/10.1073/pnas.0709698105
Kami J, Poncet V, Geffroy V, Gepts P (2006) Development of four phylogenetically-arrayed BAC libraries and sequence of the APA locus in Phaseolus vulgaris. Theor Appl Genet 112:987–998. https://doi.org/10.1007/s00122-005-0201-2
Kang YJ, Kim SK, Kim MY, Lestari P, Kim KH, Ha BK, Jun TH, Hwang WJ, Lee T, Lee J, Shim S, Yoon MY, Jang YE, Han KS, Taeprayoon P, Yoon N, Somta P, Tanya P, Kim KS, Gwag JG, Moon JK, Lee YH, Park BS, Bombarely A, Doyle JJ, Jackson SA, Schafleitner R, Srinives P, Varshney RK, Lee SH (2014) Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun 5:5443–5452. https://doi.org/10.1038/ncomms6443
Kang YJ, Satyawan D, Shim S, Lee T, Lee J, Hwang WJ, Kim SK, Lestari P, Laosatit K, Kim KH, Ha TJ, Chitikineni A, Kim MY, Ko JM, Gwag JG, Moon JK, Lee YH, Park BS, Varshney RK, Lee SH (2015) Draft genome sequence of adzuki bean, Vigna angularis. Sci Rep 5:8069–8077. https://doi.org/10.1038/srep08069
Li H, Wang W, Lin L, Zhu X, Li J, Zhu X, Chen Z (2013) Diversification of the phaseoloid legumes: effects of climate change, range expansion and habit shift. Front Plant Sci 4:386. https://doi.org/10.3389/fpls.2013.00386
Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo MC, Alhakami H, Ounit R, Hasan AM, Verdier J, Roberts PA, Santos JRP, Ndeve A, Doležel J, Vrána J, Hokin SA, Farmer AD, Cannon SB, Close TJ (2019) The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98:767–782. https://doi.org/10.1111/tpj.14349
LPWG (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. The Legume Phylogeny Working Group (LPWG). Taxon 66:44–77. https://doi.org/10.12705/661.3
Maréchal R, Mascherpa JM, Stainier F (1978) Étude taxonomique d’un groupe complexe d’espèces de genres Phaseolus et Vigna (Papilionaceae) sur la base de donneés morphologiques et polliniques, traitées par l’analyse informatique. Conservatoire et jardin botaniques, Genève. Boissiera 28:1–273
Mercado-Ruaro P, Delgado-Salinas A (1996) Karyological studies in several Mexican species of Phaseolus L. and Vigna Savi (Phaseolinae, Fabaceae). In: Pickersgill B, Lock JM (eds) Advances in legumes systematics. 8: legumes of economic importance. Royal Botanic Gardens, Kew, pp 83–87
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9. https://doi.org/10.2307/2446547
Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D (1999) Quantitative karyotyping and dual-color FISH mapping of 5S and 18S-25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42:1224–1233. https://doi.org/10.1139/g99-070
Muchero W, Diopb NN, Bhat PR et al (2009) A consensus genetic map of cowpea [Vigna unguiculata (L.) Walp] and synteny based on EST-derived SNPs. PNAS 106:18159–18164. https://doi.org/10.1073/pnas.0905886106
Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo MC, Alhakami H, Alpert M, Atokple I, Batieno BJ, Boukar O, Bozdag S, Cisse N, Drabo I, Ehlers JD, Farmer A, Fatokun C, Gu YQ, Guo YN, Huynh BL, Jackson SA, Kusi F, Lawley CT, Lucas MR, Ma Y, Timko MP, Wu J, You F, Barkley NA, Roberts PA, Lonardi S, Close TJ (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J 89:1042–1054. https://doi.org/10.1111/tpj.13404
Ouédraogo JT, Gowda BS, Jean M, Close TJ, Ehlers JD, Hall AE, Gillaspie AG, Roberts PA, Ismail AM, Bruening G, Gepts P, Timko MP, Belzile FJ (2002) An improved genetic linkage map for cowpea (Vigna unguiculata L.) combining AFLP, RFLP, RAPD, biochemical markers, and biological resistance traits. Genome 45:175–188. https://doi.org/10.1139/g01-102
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa-Harand A, Kami J, Geffroy V, Gepts P, Schweizer D (2009) Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosom Res 17:405–417. https://doi.org/10.1007/s10577-009-9031-4
Roa F, Guerra M (2012) Non-random distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet Genome Res 146(3):243–249. https://doi.org/10.1159/000440930
Sakai H, Naito K, Ogiso-Tanaka E, Takahashi Y, Iseki K, Muto C, Satou K, Teruya K, Shiroma A, Shimoji M, Hirano T, Itoh T, Kaga A, Tomooka N (2015) The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci Rep 5:16780. https://doi.org/10.1038/srep16780
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nature 46:707–713. https://doi.org/10.1038/ng.3008
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ hybridization, 1st edn. BIOS Scientific, Oxford, pp 1–203
She CW, Jiang XH, Ou LJ, Liu J, Long KL, Zhang LH, Duan WT, Zhao W, Hu JC (2015) Molecular cytogenetic characterisation and phylogenetic analysis of the seven cultivated Vigna species (Fabaceae). Plant Biol 17:268–280. https://doi.org/10.1111/plb.12174
Singh BB (2005) Cowpea [Vigna unguiculata (L.) Walp]. In: Singh RJ, Jauhar PP (eds) Genetic resources. Chromosome Engineering and Crop Improvement. CRC Press, Boca Raton, pp 117–162
Takahashi Y, Somta P, Muto C, Iseki K, Naito K, Pandiyan M, Natesan S, Tomooka N (2016) Novel genetic resources in the genus Vigna unveiled from Gene Bank accessions. PLoS One 11:e0147568. https://doi.org/10.1371/journal.pone.0147568
The Plant List (2013) Version 1.1. Available: http://www.theplantlist.org/tpl1.1/search?q=Vigna; http://www.theplantlist.org/tpl1.1/search?q=Phaseolus. Accessed 28 November 2019
Tomooka N, Vaughan DA, Moss H, Maxted N (2002) The Asian Vigna: genus Vigna subgenus Ceratotropis genetic resources. Kluwer Academic Publishers, Dordrecht, pp 1–270
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Vasconcelos EV, Fonsêca AFA, Pedrosa-Harand A, Bortoleti KCA, Benko-Iseppon AM, Costa AF, Brasileiro-Vidal AC (2015) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosom Res 23:253–266. https://doi.org/10.1007/s10577-014-9464-2
Venora G, Blangiforti S, Cremonini R (1999) Karyotype analysis of twelve species belonging to genus Vigna. Cytologia 64:117–127. https://doi.org/10.1508/cytologia.64.117
Wanzenböck EM, Schöfer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11:1007–1016. https://doi.org/10.1046/j.1365-313x.1997.11051007.x
Yang K, Tian Z, Chen C, Luo L, Zhao B, Wang Z, Yu L, Li Y, Sun Y, Li W, Chen Y, Li Y, Zhang Y, Ai D, Zhao J, Shang C, Ma Y, Wu B, Wang M, Gao L, Sun D, Zhang P, Guo F, Wang W, Li Y, Wang J, Varshney RK, Wang J, Ling HQ, Wan P (2015) Genome sequencing of adzuki bean (Vigna angularis) provides insight into high starch and low fat accumulation and domestication. PNAS 112:13213–13218. https://doi.org/10.1073/pnas.1420949112
Yundaeng C, Somta P, Amkul K, Kongjaimun A, Kaga A, Tomooka N (2019) Construction of genetic linkage map and genome dissection of domestication-related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas. Mol Gen Genomics 294:621–635. https://doi.org/10.1007/s00438-019-01536-0
Zwick MS, Hanson RR, Mcknight TD et al (1997) A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 40:138–142 https://legumeinfo.org/genomes/gbrowse/vigun.IT97K-499-35. Accessed 12 December 2019 http://rydberg.biology.colostate.edu/MicroMeasure/. Accessed 12 December 2019
Acknowledgments
The authors thank Embrapa Meio-Norte (Teresina, Brazil), Embrapa Arroz e Feijão (Santo Antônio de Goiás, Brazil), and IPK (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for supplying the seeds; Paul Gepts (University of California, Davis, United States) for supplying the BAC clones from P. vulgaris; and Valérie Geffroy (Université Paris-Sud, Orsay Cedex, France) for the bacteriophage SJ19.12 from P. vulgaris.
Funding
This study received financial support and fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Pessoal de Nível Superior: Finance Code001; InterSys Network, BioComputational Program), and FACEPE (Fundação de Amparo a Ciência e Tecnologia do Estado de Pernambuco).
Author information
Authors and Affiliations
Contributions
ARSO: performed BAC-FISH, constructed comparative maps and figures, and wrote the manuscript. LVM: performed BAC-FISH and helped to write the manuscript. FOB: helped to carry out the experiments and to write the manuscript. APH: doctorate co-supervisor of ARSO, discussed the experiments and results, maintained and provided P. vulgaris BAC clones. MMA: maintained and provided V. unguiculata BAC clones. TC: maintained and provided V. unguiculata BAC clones. AMBI: doctorate co-supervisor of ARSO and discussed the results. AFC: performed seed multiplication. ACBV: doctorate supervisor of ARSO, designed and directed the research, and corrected the manuscript. All authors read, discussed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Andreas Houben
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
Vigna aconitifolia, V. unguiculata, and Phaseolus vulgaris karyotypes share partial macrosynteny, with evidences of major chromosomal rearrangements (inversions, duplications and translocations) related to the Vigna and Phaseolus (Phaseolineae subtribe) diversification.
Electronic supplementary material
ESM 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Oliveira, A.R.d.S., Martins, L.d., Bustamante, F.d.O. et al. Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosome Res 28, 293–306 (2020). https://doi.org/10.1007/s10577-020-09635-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-020-09635-0